The FMR-VMR coregistration sub-workflow aligns functional FMR data to anatomical intra-session VMR data sets. The resulting initial alignment (IA) and fine-tuning adjustment (FA) transformation files are necessary to run subsequent FMR normalization workflows that transforms functional runs into VTC files in a target space used in the VMR normalizationthe sub-workflow. In order to specify the input and parameters of the coregistration workflow, the FMR-VMR Coregistration selection box need to be checked in the Preprocessing Workflows pane of the Preprocessing Workflow Definition.
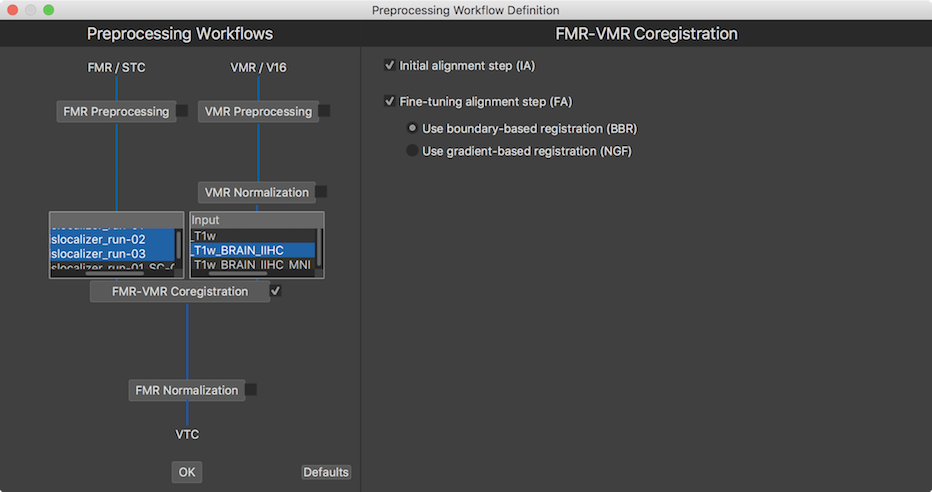
After checking the selection box of the FMR-VMR Coregistration workflow, two input selection boxes are displayed (see snapshot above), one box for selecting one (or more) FMR documents on the left side, and one selection box on the right side to select the VMR document to which the selected FMR doucments should be aligned to. In the snapshot above three runs are selected since all three functional runs of the example experiment should be aligned to the same VMR data set (for each subject). For the target VMR document, the preprocessed (brain extracted and intensity inhomogeneity corrected) VMR should be selected ending with the substring "_BRAIN_IIHC".
In the FMR-VMR Coregistration pane on the right side, settings for initial and fine-tuning adjustment can be made. The Initial alignment step (IA) option is on as default and can not be deselected since it is required as a minimum for FMR-VMR coregistration. The Fine-tuning alignment step option is also on as default but can be excluded. This is, however, not recommende dsince the fine-tuning alignment step improves functional-anatomical coregistration by correcting for small head movements that might have occured between the anatomical and functional scans. Two coregistration approaches are provided: gradient-based alignment and boundary-based registration (BBR) that can be chosen by either selecting the Use boundary-based registration (BBR) option (new since BV v20.2) or the Use gradient-based registration (NGF) option. The boundary-based registration procedure takes more time than the gradient-based registration since it involves automatic segmentation of the selected anatomical VMR to obtain a white/grey matter cortex mesh but it may produce in some cases even better results than the well-perfomring gradient-based coregistration. In most cases, however, both approaches seem to produce similar results. In case that gradient-based alignment is selected, two additional options are shown allowing to specify as the functional source either the AMR file used to visualize FMR data or the first volume of the selected functional data itself. Since BBR always uses the functional data itself, this selection is only shown when selecting gradient-based alignment.
Note. For BBR it is not necessary (and not recommended) to align all runs of a session to the same functional run during motion correction (across-run intrasession motion correction). In case that this has been chosen for gradient-based alignment, it is important to select the preprocessed FMR files as input in the FMR Input list (see section "Implications For FMR-VMR Coregistration" in topic Motion Detection and Correction). For BBR it is recommended ot select the unpreprocessed FMR documents as input; as long as the FMR data is not intensively smoothed, the preprocessed FMR documents can, however, also be used when chosing the BBR approach for FMR-VMR coregistration.
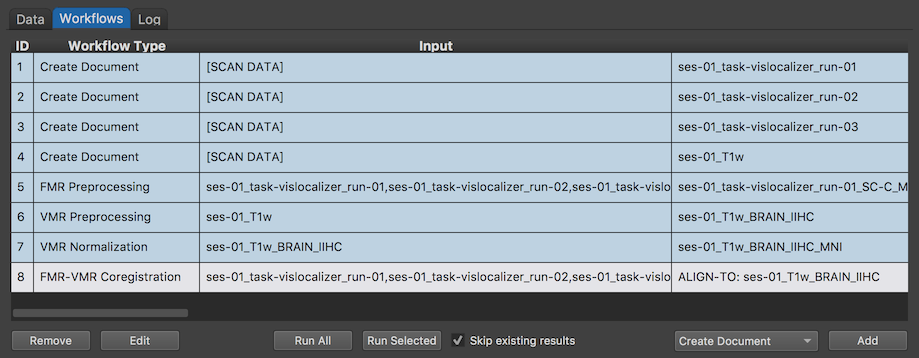
After clicking the OK button, the specified FMR-VMR coregistration workflow appears in the Worfklows tab (see row 8 in the Workflows table shown above). To execute it, the Run Selected button can be clicked after selecting the coregistration workflow in the table. The Log tab will report the progress of the workflow involving in this case the coregistration of 6 FMR documents (3 runs are aligned to the respective VMR document for 2 subjects). This workflow will not result in entries in the subject-specific data tables which display main BrainVoyager documents only but the resulting "_IA.trf" (for the initial alignment step) and "_FA.trf" (for the fine-tuning alignment step) files will be stored in the created workflow folder for each subject ("workflow_id-8_type-4_name-functoanatcoreg").
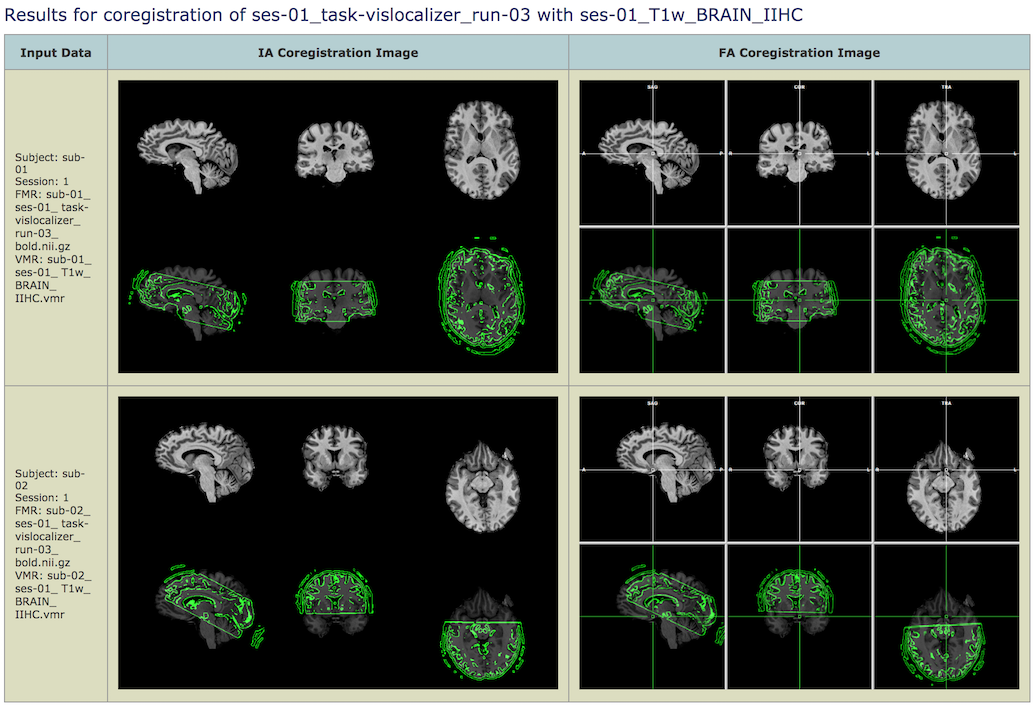
The screenshot above shows part of the produced workflow report of the example data (coregistration result of third run for subject 1 and 2) after using gradient-based fine-tuning alignment. This visual report is very helpful in order to check whether FMR-VMR coregistration has worked correctly in all runs for all subjects. To find sources of potential issues, the report shows both the result of the initial alignment (IA) step (column IA Coregistration Image on the left) as well as the result of the fine-tuning adjustment (FA) step if included when defining the workflow (column FA Coregistration Image on the right). The screenshot below shows the workflow report when using BBR for the fine-tuning adjustment step.
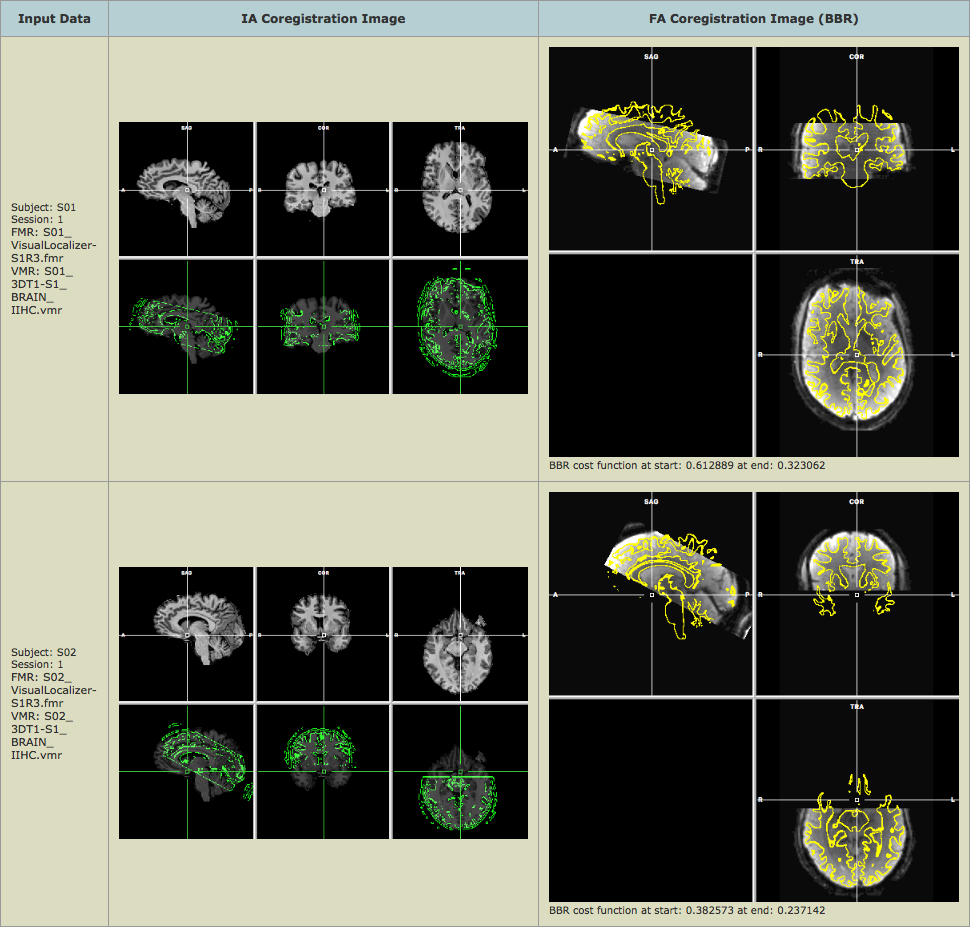
In case of BBR, the images for the fine-tuning adjustment report right side) look different than in the gradient-based FA report since they show the quality of alignment by projecting the vertices of the reconstructed white-grey matter boundary mesh of the segmented anatomical intra-session data set into the first functional volume of the respective FMR data (stored in a VMR after running the initial alignment step). The achieved alignment accuracy is also displayed numerically below the FA image in the text string "BBR cost function at start: [value 1] at end: [value 2]". Smaller values of the displayed cost function are better and values below 0.5 indicate very good alignment. The values 0.32 (subject 1, run 3) and 0.23 (subject 2, run 3) thus indicate successful FMR-VMR coregistration; furthermore the displayed values indicate that BBR improved alignment substantially beyond header-based alignment (cost function decreased from 0.61 to 0.32 for subject 1 and from 0.38 to 0.23 for subject 2).