Turbo-BrainVoyager v4.4
Output Generation
The Output tab of the Data Simulator is used to actually generate simulated data according to the settings specified in the other tabs by clicking the Generate Data button. Note that any changes made in the dialog will automatically be saved to the current SIMJ file when clicking on Generate Data, except if one turns off the Save settings option.
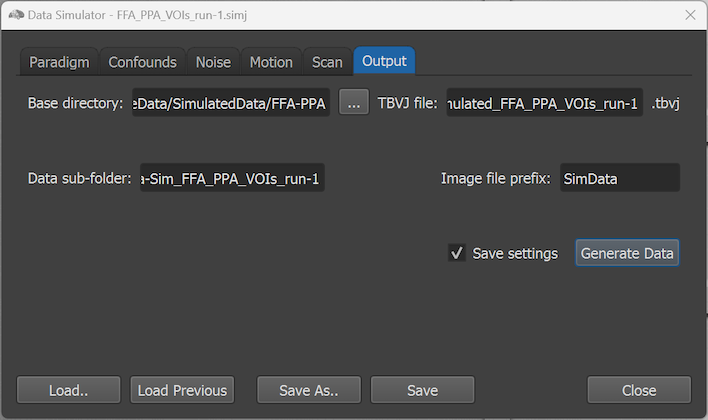
The path to the current SIMJ file appears in the Base directory text field above. It is possible to change the default base directory using the Browse Base Directory button on the right side of the Base directory field. After clicking the Generate Data button, all generated files will be stored within a subfolder of the base directory, which will be named:
[base_directory]/Generated_[simj-base-name]
This template string contains variable names in square brackets that are filled with concrete names when data is generated. The string above refers to the [base directory] and the [simj-base-name], which resolves to the current file name without the '.simj' extension as well as without any run specifier (e.g., _run-1.simj will be removed for the SIMJ file used in the example). Removing the 'run' specifier is important since it allows to store multiple simulated runs belonging to the same experiment in the same 'Generated' folder. Since in the example case the SIMJ file is named
FFA_PPA_VOIs_run-1.simj
all data will be saved in a sub-folder with the name:
[base_directory]/Geerated_FFA_PPA_VOIs
The generate data in that folder includes not only the functional volumes but also a TBVJ file that is needed, as usual, to control real-time data analysis. Furthermore, provided protocol and region files as well as anatomical brain data will be stored on disk. The main generated files are the functional data, which are stored as a series of 3D NIfTI files in the data sub-folder that is shown in the Data sub-folder text field (see screenshot above). The default name of the data sub-folder is generated according to the following template:
Data-Sim_[simj-name]
The 'simj-name' here includes the run specifier but not the extension, i.e. in the example case the data sub-folder name is:
Data-Sim_FFA_PPA_VOIs_run-1
The individual 3D functional volumes are stored using the prefix obtained from the Image file prefix text field (see screenshot above), which is 'SimData' as default. After the prefix, two fields ('e' for echo, always 1 in generated data, 'd' for data, a counter of volumes) are appended according to the scheme described in topic Settings for Philips NIfTI Data:
SimData-e1-d1.nii
SimData_e1_d2.nii
SimData_e1_d3.nii
:
:
Note that the NIfTI volumes are saved uncompressed for fast processing during generation and later real-time analysis.
While the functional NIfTI files contain EPI-like brain slices (if enabled in the Scan tab), a 3D T1-weighted dataset (CG_T1w_IIHC_MNI.vmr) is stored in the Anatomy directory inside the Generated folder.
The generated TBVJ file is placed in the 'Generated' folder under the name:
Simulated_[simj-name].tbvj
The TBVJ file is loaded automatically, and real-time processing can, thus, be started immediately by clicking the red Start button in the main window of TBV. Depending on settings, there are also some additional files stored in the Generated folder, including a '.mot' file containing the values of simulated motion effects over time. This file may be useful to compare it with the detected (and corrected) motion parameters during real-time processing of the data.
In summary, the directory tree after generating the simulated data is as follows:
- [base_directory]
[simj_file]
[prt_file] (here or elsewhere)
[voi/roi_file] (here or elsewhere)
- [Generated_[simj-base-name]
Simulated_[simj-name[.tbvj
Simulated_[simj-name[.mot (if motion simulated)
[simj-name].rcps (useful if patterns simulated)
- Anatomy
CG_T1w_IIHC_MNI.vmr
CG_T1w_IIHC_TO_MNI_a12.trf
- Data-Sim_[simj-name]
SimData-e1-d1.nii
SimData_e1_d2.nii
SimData_e1_d3.nii
:
:
The screenshot below has been made in the middle of real-time analysis of the dataset generated by the example SIMJ file. Note the time courses for the simulated FFA activation effects (blue ROI) and PPA in the right hemisphere (shown on left side of EPI-like slices) exhibiting opposite preferences as expected from the settings made in the Paradigm tab.
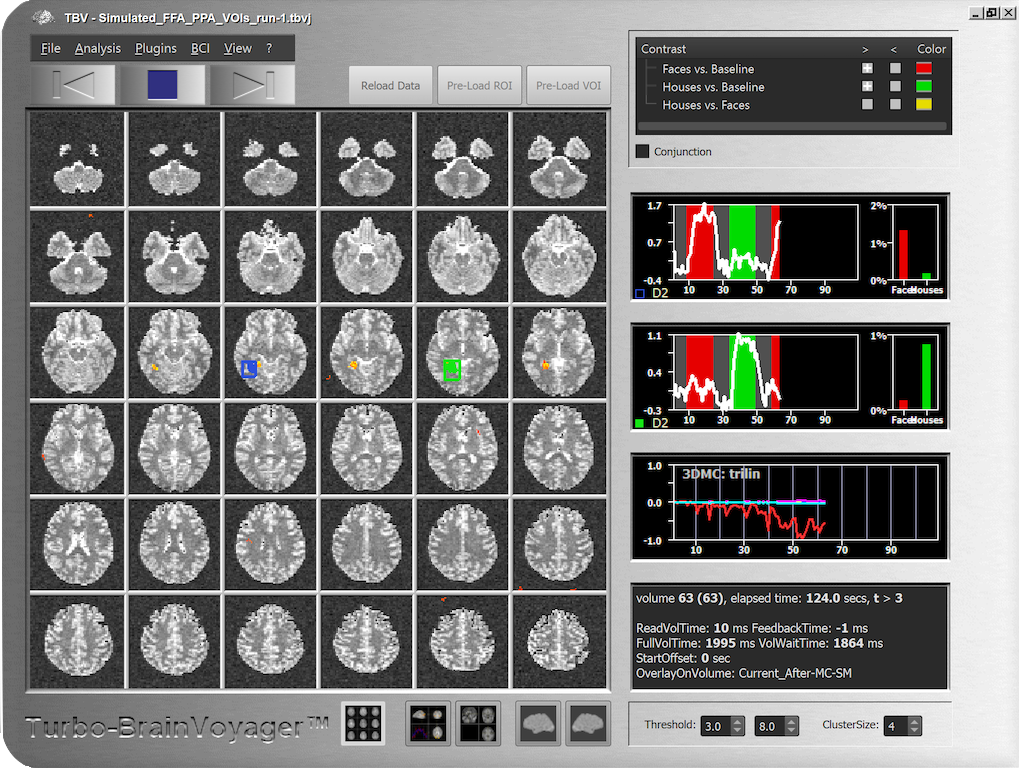
Also note the motion correction window, which shows only detected motion along the X axis (red curve) but not other axes as specified in the Motion tab. It is now very easy to go back to the Data Simulator and change parameters such as the amount of noise or motion to see the corresponding results in the real-time analysis. Note also that you can modify the generated TBV file by opening the TBV Settings dialog to change processing options such as drift removal or turning off motion correction.
Copyright © 2002 - 2024 Rainer Goebel. All rights reserved.