Turbo-BrainVoyager v4.4
Overview
Performing real-time analysis with Turbo-BrainVoyager requires specification of a TBV settings file containing information about the upcoming functional measurements. If a proper TBV settings file has been prepared - usually with the help of the TBV Settings dialog - real-time analysis can be started by clicking the red Start button. After analysis has started, statistical maps will appear in various visualizations, including multi-slice and volumetric displays. Using the mouse, any region can be selected to show incrementally growing time courses. The extracted time course of regions can also be used for BCI applications such as neurofeedback. In order to learn how to use these and many other features of Turbo-BrainVoyager, it is recommended to run one of the available example data sets.
Preparing Real-Time fMRI Analysis
A TBV settings file can be easily created and modified using the TBV Settings dialog. Once a settings file has been created matching a particular scanner environment and data format, only small adjustments are typically necessary to create TBV settings files for later experiments at the same scanner. To open a settings file from disk use the Open TBV File item in the File menu and select an existing ".tbvj" file, which will automatically be presented in the TBV Settings dialog. In order to modify an existing TBV settings file, use the Edit TBV File item in the File menu. When launching Turbo-BrainVoyager the next time, it automatically loads the last used TBV settings file. This allows to inspect and re-analyze the data of the last experiment by clicking the Reload Data button (in case that the generated data at the end of a real-time run has not been removed). In case that the next functional run uses the same settings as the previous one, only a few changes need to be made before one can click the Record main button for the next run. The changes that are likely required are updating the "series" number of the scan, the folder containing the new data (if changes on the scanner side) and eventually a new experimental protocol in case that the next run uses different conditions or temporal order of conditions. With these changes made, a new TBV file should be prepared by using the Save As button in the TBV Settings dialog. Note that the TBV settings files for all planned functional runs of a real-time scanning session (e.g. with different protocl files) can be prepared before the actual scanning session starts reducing the work to be done between successive runs.
Running Real-Time fMRI Study
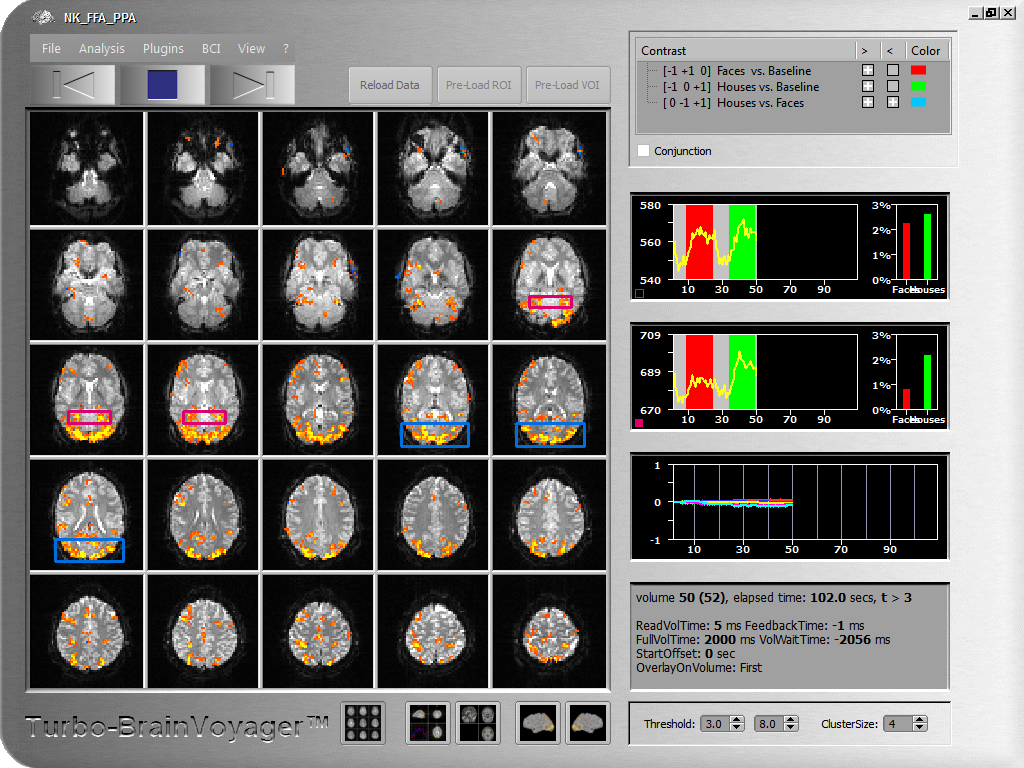
As soon as data becomes available from the scanner, TBV reads it volume-by-volume from a folder, called the WatchFolder in the TBV Settings dialog. In case not already done by the scanner, a few volumes at the begin of the run may be discarded by setting the NrOfSkippedVolumes entry in the settings file to avoid T1 saturation effects (high intensity values) at the begin of a functional run. If specified, 3D motion correction is performed aligning each (but the first) volume to a reference volume, which is either the first volume of the current run or the first volume of a previous run. The detected 6 motion parameters are plotted in a time course window. Each motion corrected volume may be spatially smoothed to increase signal to noise. Incoming volumes (time points) are incrementally statistically analyzed using a General Linear Model (recursive least squares GLM) based on an automatically created design matrix using the provided stimulation protocol. Hypothesis testing is performed using contrasts, which are automatically created but can also be specified explicitly using a ContrastFile containing any desired contrast definitions. Available contrasts are shown in the contrast list at the right upper corner of the main window and may be turned on and off for display at any time during real-time analysis. Up to 4 contrasts may be displayed simultaneously (out of arbitrary many contrasts), each with its own color or with a generic look-up table. Additional confound predictors may be added to remove linear trends and non-linear (low-frequency) drifts. It is also possible to add detected motion parameters incrementally to the design matrix for removal of drifts and motion-related noise. The incrementally calculated contrast t maps are thresholded with a value specified in the TBV settings file in combination with a cluster size value. The statistical threshold can also be adjusted on the fly during analysis The thresholded maps are displayed on measured slices in the default Multi-Slice View as well as on more advanced (normalized) anatomical and cortical surface-based visualizations (see below).
Advanced Brain Visualization Options
Various brain visualizations are available. The default visualization is a multi-slice view providing a good overview of the updated statistical information on top of the original slices. In single slice view, spatial details and information from individual voxels may be obtained. In a 3D view, the original data may be viewed on 3 slice planes (sagittal, coronal, axial). This orthographic 3D view may also show high-resolution anatomical information if available (or created) prior to the functional scan. After proper preprocessing, the functional data may run as a movie on high-resolution data in MNI, AC-PC or Talairach space as well as on cortex mesh visualizations. To inspect the time course of individual voxels or regions-of-interest, various tools are available to easily select interesting ROIs from any of the described visualizations. Selected raw or detrended time courses are shown in time course windows together with incrementally updated bar plots showing estimated beta values for main conditions. An event-related averaging plot for the current ROI is visualized in one quadrant of the orthographic 3D view visualizations.
Copyright © 2002 - 2024 Rainer Goebel. All rights reserved.