BrainVoyager v23.0
4D NIfTI Time Course Files
In case that a loaded NIfTI file contains 4D data, BrainVoyager assumes that the file's content represents fMRI time course data (if you want to load a stack of maps, use the Volume Maps dialog).
From Scanner Space to FMR-STC Documents
In case that the 4D time course data refers to a "Scanner" coordinate system (qform or sform code: 1), the 4D data is converted to a (native space) BrainVoyager FMR-STC document. In case of normalized (e.g. MNI-152) space, the file is converted in a BrainVoyager VTC document. This works at present only, if the transformation matrix does not contain rotations. The created STC or VTC data are stored as float values even if the data type in the original NIfTI file are stored in a different format.
As described above, the NIfTI qform (or sform) transformation matrix is not applied in case of Scanner space (STC) data but it is stored in the FMR header as a (past) spatial transformation (see below). There will be also no to-BV axes reorientation applied, i.e the displayed slices are shown as stored in the NIfTI file. The snapshot below shows an FMR-STC document resulting from opening a 4D NIfTI file that has been creaed from the "Objects" sample data by converting the DICOM files to a NIfTI 4D file.
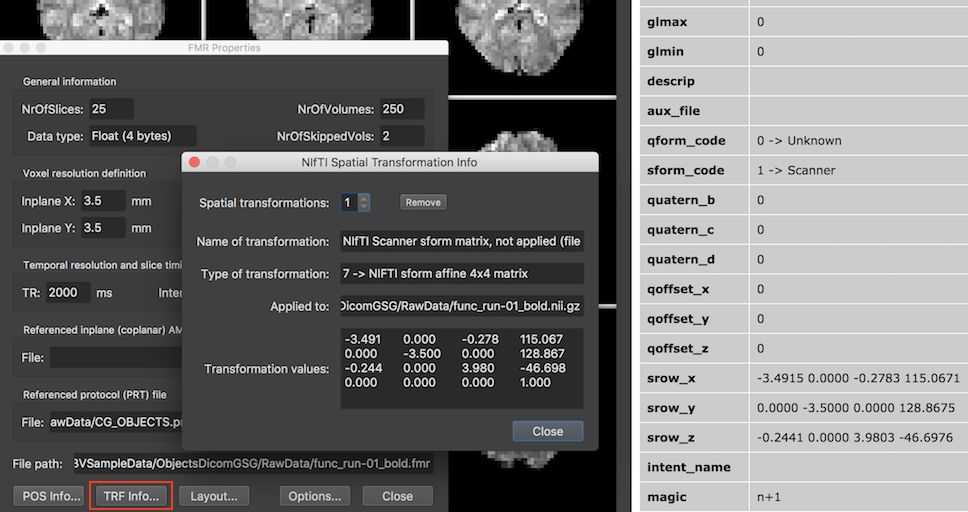
Note that the standard FMR Properties dialog displays a TRF Info button (see red rectangle) in case that the FMR header contains special spatial transformation information, otherwise the button is usually not shown. Clicking on this button shows the NIfTI Spatial Transformation Info dialog displaying the stored NIfTI orientation information (see screenshot above). The FMR header will contain correct voxel size values extracted from the NIfTI header allowing to run spatial preprocessing steps such as motion correction and spatial smoothing. The TR should also be correct (if set in the pixdim field for the 4th dimension) but except if provided in a sidecar file, the Inter Slice Time might be missing. To be on the safe side, it is recommended for temporal preprocessing (drift removal, temporal smoothing) to use values referring to time points and not with respect to seconds. As long as no multi-band sequence is used, a code for the slice ordering will be extracted from the NIfTI header if available but slice scan time correction might not be run correctly in case that a multi-band (simultaneous multi-slice) sequence has been used. When the 4D data was created by BrainVoyager, its DICOM-to-NIfTI converter stores all necessary extra information in an associated JSON sidecar file, including the SIEMENS slice timing table. In combination with the JSON sidecar file, 4D time course NIfTI files can be used as full replacements for FMR-STC (and VMR-VTC) files.
From Normalized Space to VMR-VTC Documents
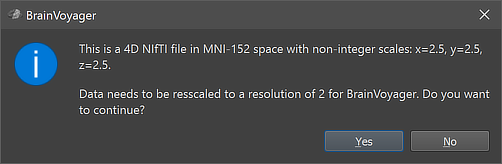
Since BrainVoyager 22.2 4D NIfTI files in normalized space can be opened using the Open NIfTI icon in the main toolbar or the respective item in the File menu. At present, the data is converted to a VTC file in case that the sform transformation matrix does not contain rotations, which is usually the case. Most transformation matrices contain however scaling values. In case that they are integral (e.g. 2.0, 3.0), data will not be resampled when converting to a VTC document, which is the case, for example, when loading VTC data that had been saved as NIfTI using BrainVoyager. When importing normalized 4D data that has been generated by other software, scaling values will often be non-integral. In that case a pop-up window will appear (see screenshot above) that explains that the data will be resampled to be compatible with BrainVoyager's standard handling of VTC files. In the example above, the functional voxels have a size of 2.5 millimeter and the dialog states that the data will be resampled (oversampled) to 2.0 mm when proceeding. The screenshot below shows the resulting VMR-VTC document as well as part of the 4D NIfTI header.
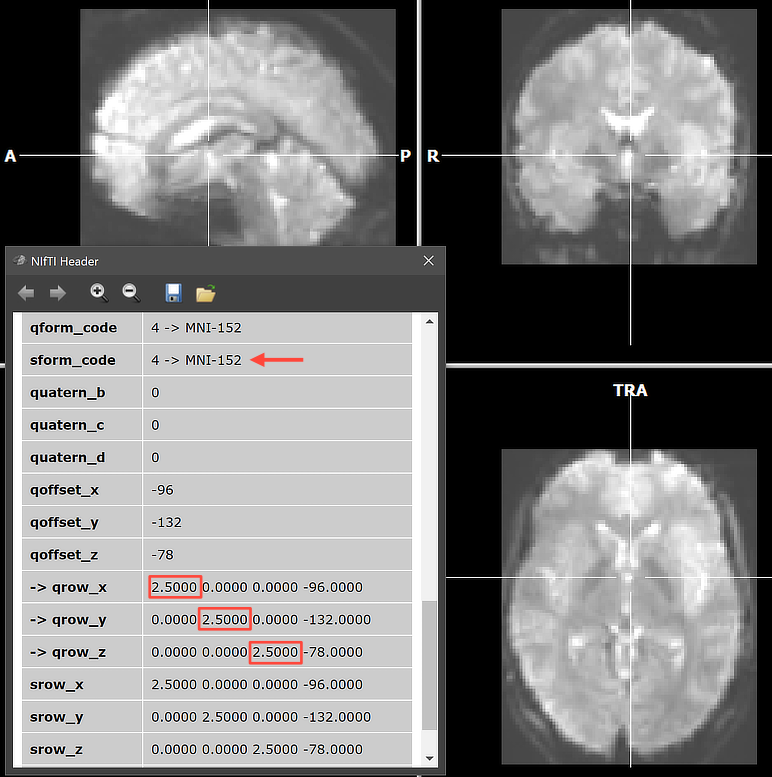
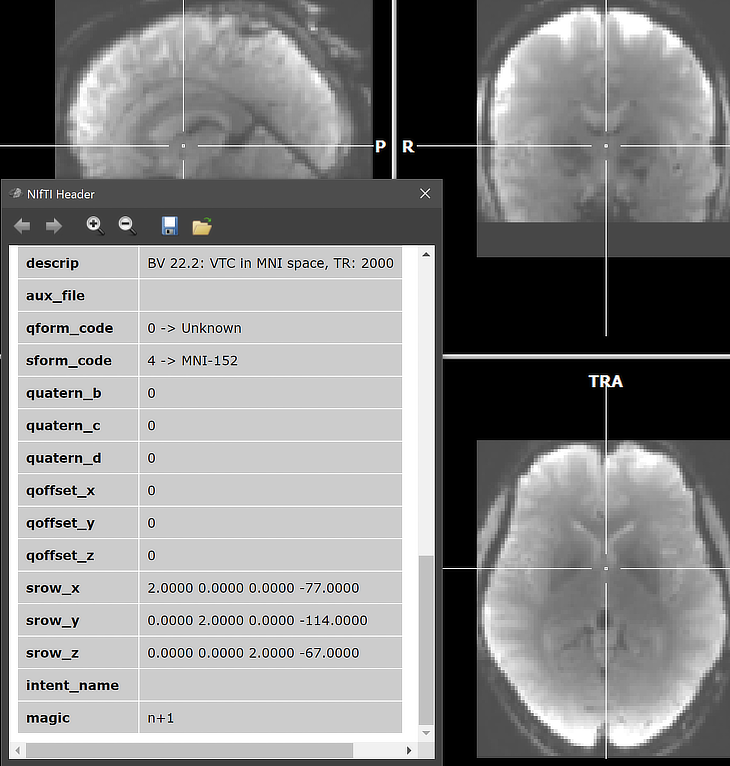
To show the resulting VTC data, its first volume is converted into a VMR document and used to host the converted VTC data. One can, of course load any relevant (T1 weighted) anatomical document and then link the saved VTC file as usual. Note that in case a NIfTI .TSV sidecar protocol file wth the same base name is found, it will be converted into a BrainVoyager protocol. In that case, it is, for example, possible to perform a single-run GLM analysis directly after opening a 4D NIfTI file. Note that for normalized data, the functional volumes are placed in the standard BrainVoyager bounding box. Since BrainVoyager 22.2, the MNI bounding box has been slightly extended and is defined (in mm) along the X (left-right) axis as -75 to 75, along the Y (posterior-anterior) axis as -115 to 77, and along the Z (inferior-superior) axis as -68 to 88. These values can be adjusted to custom values in the Data tab of the Settings dialog. Note that it is recommended to keep the same bounding box values for all VTC files of the same project, no matter whether they are imported from NIfTI files or generated with BrainVoyager;s Create VTC dialog.
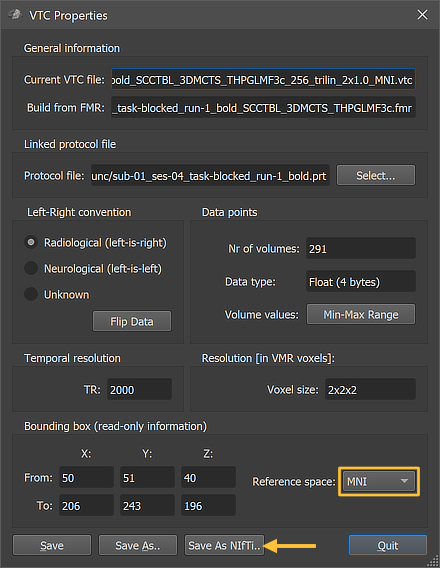
Normalized (e.g. MNI space) VTC files can be exported as 4D NIfTI files using the Save As NIfTI button in the VTC Properties dialog (see arrow above). The screenshot below shows the (re-)imported VMR-VTC data and part of the NIfTI header that had been generated by BrainVoyager when the VTC file had been exported as NIfTI. The 'descrip' string of the NIfTI header indicates that this file was produced by BrainVoyager 22.2. Note also that the scaling values are integral (2.0) as expected.
Copyright © 2023 Rainer Goebel. All rights reserved.