BrainVoyager v23.0
NIfTI Volume Map Files
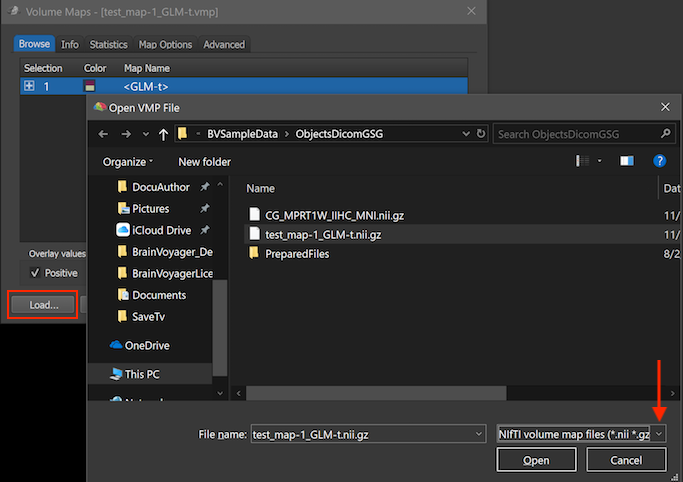
BrainVoyager supports (since version 21.2) direct loading and saving of NIfTI volume maps using the Load, Add and Save buttons in the Volume Maps dialog. When opening or saving volume map data, the appearing Open File or Save File dialog offers two file type options, one for standard BrainVoyager volume map (VMP) files and one for NIfTI volume map files. The (Windows 10) screenshot below shows the Volume Maps dialog on the left and the Open VMP File dialog on the right that appears after clicking the Load button (see red rectangle) in the Volume Maps dialog. Using the selection box in the right lower area (see red arrow), either the Volume map files (VMPs) or the NIfTI volume map files file type can be chosen for selecting a file on disk. In a similar way either the standard volume map type or the NIfTI file type can be selected when adding maps or when saving volume map data to disk.
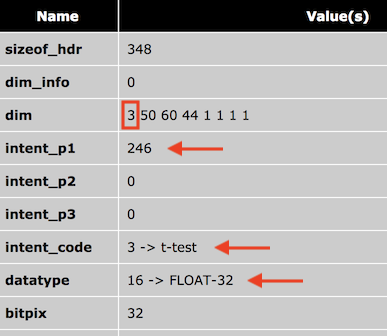
A NIfTI file is identified as a (statistical) map if its header specifies a 3D or 4D data volume containing float or double values (see screenshot below). The header also contains an intent code that, if filled, specifies the stored (statistical) data, e.g. whether the map contains t test, F test, correlation or z score values (among others). Depending on the stored statistical data, additional parameters (up to 3) may be stored in additional intent parameter fields such as the degrees of freedom of the respective statistical tests. In the screenshot below, the intent code "3" specifies that the stored data contains t test values and the intent parameter 1 (see "intent_p1" entry) contains the degrees-of-freedom of the t test. The NIfTI standard also allows to store different parameter values per voxel but this possibility is not (yet) supported by BrainVoyager.
BrainVoyager supports saving volume maps in NIfTI format if they are represented in normalized (MNI-152, Talairach) space as well as in ACPC and native space (both coded as "Aligned" space in the NIfTI header). In the case of MNI and Talairach space data, it is ensured that the exported NIfTI volume map files will be interpreted correctly by other neuroimaging software. The exported NIfTI volume map files can thus be shared with colleagues and uploaded to (public) databases expecting analysis results as NIfTI float maps. Note that a normalized anatomy file can also be provided in NIfTI format by saving the normalixed VMR (e.g. single subject or averaged group anatomy in MNI-152 or Talairach space) to disk using the Save NIfTI item in the File menu. If a volume map in ACPC space is stored as a NIfTI file, it is marked as being in "Aligned" space since the NIfTI format does not store a specific value for ACPC space in the s(q)form_code. BrainVoyager puts the space in the description text field (see below) to convey this information. The "Aligned" space is also used when saving NIfTI volume maps but these files will probably be useful only for re-reading them in BrainVoyager at a later time. Also note that some fields in the NIfTI header are used in a specific way to support extended information used by BrainVoyager. The description field of the NIfTI header, for example, starts with a string that identifies BrainVoyager as the creator of the file as well as the used BrainVoyager version (e.g. "BV 21.2") followed by additional information (see below).
Note. The NIfTI header is not shown when loading a volume map file from the Volume Maps dialog. In order to view the NIfTI header of a volume map file, it need to be loaded using the Open NIfTI icon or Open NIfTI item in the File menu. Since a volume map file needs a hosting VMR file in BrainVoyager, the selected NIfTI volume map file will be loaded both as a VMR file as well as a map file which is overlaid on top of the (pseudo) VMR data.
Saving a NIfTI Volume Map
When exporting VMP data containing multiple maps, each sub-map is exported as a separate 3D NIfTI volume map file. BrainVoyager suggests the current (VMP) file name as the core name of the to-be-stored NIfTI file appending the name of the respective sub-map. A map called "Images" in the VMP file "GLM-Contrasts.vmp" will be stored (if suggested name is not changed) with the file name "GLM-Contrasts_map-1_Images.nii.gz". A second map "Sounds" will be stored as "GLM-Contrasts_map-2_Sounds.nii.gz". The "map-[n]" substring ensures that exported NIfTI files obtain a unique file name even in case that the map names are identical.
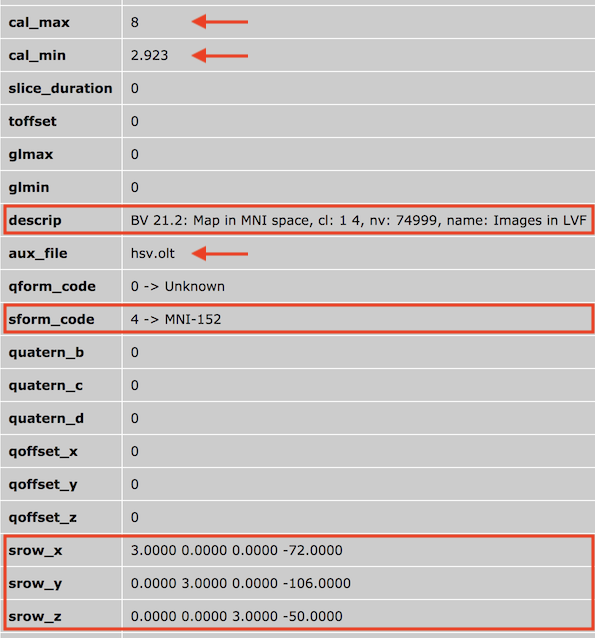
Saving volume maps as NIfTI file(s) does not resample the VMP float values, i.e. the map 3D "cube" is stored without changing the values; the BrainVoyager native orientation is, however, converted into neurological RAS orientation when saving NIfTI volume map files to disk. The sform transformation matrix (describing how the voxel indices in the file are mapped into the intended space) can then be set to a scale / translation matrix without rotations. BrainVoyager uses only the sform matrix to store transformations (see "srow" fields in the screenshot above), i.e. the qform transformation entries are not used. In case the intended space is normalized (MNI, Talairach), the stored scaling values adjust the resolution of the map to 1mm, if necessary. This includes the relative resolution of the map with respect to the hosting VMR anatomy as well as the resolution of the hosting VMR in case it is not a 1mm isotropic data volume (1mm resolution is recommended). Translation values are stored in the fourth column of the sform transformation matrix describing how to shift the 3D map to the right position with respect to the intended normalized space. Note that it is advised to use the same isotropic resolution (1mm recommended) for hosting VMRs when saving and loading NIfTI map files. If the VMR resolution used when saving a NIfTI map is different from the resolution when loading a NIfTI map, the volume maps need to be resampled; this implies that the data will be treated in a generic way as if it did not come from BrainVoyager.
BrainVoyager uses the NIfTI header cal_min and cal_max fields to store the minimum and maximum threshold of the saved map (see screenshot above). Other relevant values are stored in the description (text) field, including the "Map in [referemce space] space" substring, the "cl: [flag] [value]" substring, the "nv: [value]" substring and the "name: [text]" substring. The first substring informs a user of the file about the reference space as originally assigned by BrainVoyager. For MNI-152 and Talairach space, this matches the space coded in the sform_code field in the NIfTI header. BrainVoyager's ACPC as well as Native reference space are coded as "Aligned" in the NIfTI header. The "cl" substring stores a flag that specifies whether the volume map uses a cluster threshold (value: "1") or not (value "0") followed by the cluster threshold value (number of connected map voxels). The "nv" field stores the number of map voxels with a value unequal 0.0. This number is usually less than the full number of voxels of the map "cube" and is used e.g. for FDR and Bonferroni correction. The "name" field stores the map name, which might be truncated in case that the string extends beyond the limits of the description header field (80 characters). In case that a non-default overlay look-up table (OLT) is used for the saved volume map, the OLT file name is stored in the aux_file field (see screenshot above) of the NIfTI header (truncated in case the OLT file name extends beyond the maximum of 24 characters).
Loading a NIfTI Volume Map
When a NIfTI volume map file is loaded from the Volume Maps dialog, it is checked whether the reference space of the 3D map file fits to the reference space of the hosting VMR. It is also checked whether the volume map contains real (float or double) values, other values are currently not accepted. Then the description text string of the NIfTI header is analyzed in order to check whether the map was created by BrainVoyager, i.e. whether the text starts with the "BV " substring. If this is the case, the transformation matrix stored in the srow fields is analyzed to check whether it is a scale/translation matrix without rotation. In this case the scale values are extracted and if they are integral multiples (1, 2, 3) with respect to the hosting VMR, the map is treated like a standard BrainVoyager volume map, i.e. the map is shifted to the appropriate position within the normalized reference space using the translation values in the 4th column of the transformation matrix without resampling the map values. In case the data comes from external software, the transformation matrix is applied as is including potential rotations and scales. Also in case that a BrainVoyager stored NIfTI volume map can not be loaded as is, e.g. when the hosting VMR resolution is not the same as when saving the data, the volume map is treated in the most general way including resampling the data.
In case that the volume map is identified as stored by BrainVoyager, some values stored in the header (see above) are interpreted in a specific way. In order to re-threshold the map in the same way as it was thresholded when saving the map as a NIfTI file, BrainVoyager extracts the threshold from the cal_min field of the NIfTI header. The upper "threshold" (usually not used to exclude values but to map the upper boundary of the color range of the overlay look-up table to the data values) is extracted from the cal_max value. In case that the aux_file text field contains a string ending with ".olt", it is interpreted as a standard overlay look-up table file that is loaded (if present) from the MapLUT folder, which is located in the BrainVoyager installation folder. If no OLT file is found in the aux_file text field, the default volume map look-up table is used.
Note. BrainVoyager uses the (positive) map threshold value for both positive and negative map values. Other software reading NIfTI files might interpret the stored cal_min and cal_max values as a filter, i.e. only positive values in the respective value range might be shown. In that case, the range/threshold values in other software should be adjusted, e.g. to also include negative values.
Copyright © 2023 Rainer Goebel. All rights reserved.