BrainVoyager v23.0
NIfTI Files
NIfTI files can be directly loaded from the Open NIfTI icon in the main toolbar or the Open NIfTI item in the File menu. Furthermore, NIfTI volume map files can be saved and loaded directly from the Volume Maps dialog. and 4D NIfT time course files can be saved and loaded and saved using the respective commands in the File menu and the VTC Properties dialog. NIfTII files are thus becoming as easy to use as standard BrainVoyager 3D and 4D data files including proper handling of NIfTI transformation matrices (see below). This topic describes the steps that are performed when loading anatomical (3D) NIfTI files as well as how Scanner space functional NIfTI data can be coregistered to same-session anatomical datasets. Support for (normalized) 4D data is described in topic 4D NIfTI Time Course Files, and support for (statistical) volume maps is described in topic NIfTI Volume Map Files.
NIfTi (version 1.1) files are supported as single files (.nii), compressed single files (.nii.gz) and file pairs (.hdr, .img). At present 3D (usually anatomical or statistical map) data and 4D time course data with NIfTI data type values of 2 (BYTE), 4 (INT16), 512 (UNSIGNED INT16), 8 (INT32), 16 (FLOAT32) and 64 (DOUBLE) are supported when opening NIfTI files from the File menu or main toolbar. Anatomical 3D data is converted into VMR (and V16) files with 1 byte (UINT8) and 2 bytes (UINT16) per data point. Statistical map data is converted to a volume map that is overlaid on the same data interpreted as a VMR (see topic NIfTI Volume Map Files for handling volume maps directly from the Volume Maps dialog). 4D time course data is converted into 4-byte float (FLOAT32) FMR/STC data in case the data is in Scanner ("native") space, and into 4-byte float VTC files if the data is in normalized (MNI, TAL) space. For more general NIfTI import and export options, one can use the NIFTI converter plugin, that can be invoked using the NIfTI-1 converter item in the NIfTI GIfTI Plugins sub-menu within the Plugins menu. The following descriptions focus on 3D anatomical example data but the described NIfTI-to-BV transformation steps are the same for any supported 3D/4D files. Since the performed steps are different for normalized (MNI, Talairach) and non-normalized (Scanner) space data, some information about how coordinate systems are stored in the NIfTI header is provided in the next section. For more information, consult the official description of the NIfTI-1 header.
NIfTI Coordinate Systems
The NIfTI format specifies two (out of three) methods to ensure that the orientation of the data in a NIfTI file is properly interpreted by neuroimaging software. Both methods in effect specify an affine transformation matrix that maps voxel i,j,k coordinates to RAS+ (left-to-right, posterior-to-anterior, inferior-to-superior) world coordinates. One method (method #2, qform) is based on a rotation matrix (stored as a quaternion), scaling values and a translation vector; this method is mainly used to map voxel coordinates to x,y,z coordinates in scanner world space (or to another data set for alignment). The other method (method #3, sform) uses a full 12-parameter affine matrix that maps voxel coordinates to x,y,z MNI-152 or Talairach space, which also use a RAS+ coordinate system. If a data set is already transformed in MNI or Talairach space, the sform transformation matrix is simply the identity matrix (eventually with 90/180 degree roations and axis flips to reorient coordinate axes) combined with potential scaling and translation values. A third method is available that only supports scaling the data (using the NIfTI pixdim values) without defining a coordinate system; this method is not recommended and only included in the NIfTI specification for compatibility with the older Analyze 7.5 file format.
Both affine transformation matrices of method #1 and #2 can be present in the same NIfTI file. In order to know which transformation is valid, the NIfTI header contains a value for each transformation type, the qform_code and the sform_code. If one (or both) codes contain a value > 0, the respective transformation is valid and can be used. If both codes are 0, only the non-recommended third method (using only scaling transformations) is available. The two forms were introduced so that two transformations could be defined simultaneously (e.g. one transformation to align the data to scanner space and one transformation to align the data to normalized space) but in practice most imaging software keep the two transformations identical or use only one. The non-zero qform/sform code specifies the target transformation RAS+ space as follows:
- Code = 1, "Scanner" - the transformation matrix maps voxel coordinates to scanner RAS world coordinates.
- Code = 2, "Aligned" - the transformation matrix maps voxel coordinates to some (usually subject-specific) aligned RAS space.
- Code = 3, "Talairach" - the transformation matrix maps voxel coordinates to Talairch coordinates (e.g. x=0,y=0,z=0 corresponds to the AC point).
- Code = 4, "MNI" - the transformation matrix maps voxel coordinates to MNI-152 coordinates.
From NIfTI to BV Coordinates
The "Scanner" space usually corresponds to the orientation data stored in DICOM files. When starting with DICOM data while running the standard document creation tools, BrainVoyager stores the DICOM orientation data in the VMR (and FMR) header and uses it later when aligning functional and anatomical data (initial header-based alignment); the anatomical and functional data arrays are stored and visualized in original voxel space in order to avoid resampling the data (except for integral multiples of 90 degree rotations and axis flips to reorient the data to BrainVoyager's standard "sagittal" coordinate system). The same strategy is performed now as default when reading NIfTI files with a "Scanner" transformation matrix, i.e. the original matrices are stored in the header and can be used for FMR-VMR alignment but the data are stored and presented in voxel space, i.e. coorinates are direct indices in the voxel data arrays. Since only (multiple of) 90 degree axes adjustments are performed, this transformation is called "ortho" in the Preferences dialog (see below). An important advantage of the ortho transformation is that the data needs not to be resampled (e.g. scaled or rotated). The transformation matrix for "Aligned" space data sets are also only ortho-applied (as default) when importing data while the full transformation matrix is stored in the BrainVoyager (VMR / FMR) header. When, however, a NIfTI file contains a "MNI" or "Talairach" transformation matrix, the full matrix is applied to the voxel coordinates and the data is presented in standard MNI-152 or Talairach space. While this "full" transformation may require data resampling, this approach allows direct application of standard analysis tools in BrainVoyager without manual preparation.
In summary, when reading NIfTI files with a qform (or sform) code of 1 (Scanner), BrainVoyager stores the original (qform or sform) transformation matrix in the resulting VMR (FMR) header and performs only an ortho-reorientation. The stored matrix can be used when coregistering a loaded NIfTI 4D data set (as FMR) to a VMR volume from the same run. If, however, the sform (or qform) code value is 3 (TAL) or 4 (MNI-152), BrainVoyager applies the full transformation matrix which may require resampling the data (if not stored in normalized 1 mm space). Ensuring that the full transformation matrix is applied for anatomical data in normalized space allows to link VTC data to the loaded VMR in case it has been normalized to the same space (Talairach or MNI-152). The default behavior for different NIfTI reference spaces can be changed in the Preferences / Settings dialog (see below). While the original NIfTI qform or sform is stored in the VMR (or FMR) header, it is also saved (in the VMR/FMR "past transformations" header data structure) whether an ortho, full or no transformation has been performed.
Since BrainVoyager does not use a RAS+ coordinate system internally, the ortho or full RAS+ volume is reoriented to BV's internal PIL+ (or PIR+) coordinate system (anterior-to-posterior, superior-to-inferior, right-to-left or left-to-right). Since BrainVoyager displays coordinates in LAS+ or RAS+ space, the data is presented to the user as intended by the NIfTI file (except for the default orientation of the x axis, see below). Note that 4D NIfTI files in Scanner space are imported as FMR-STC data without any coordinate transformation. The NIfTI qform (or sform) transformation matrix is, however, stored in the FMR header so that the orientation of the data is known and usable for FMR-VMR coregistration.
Opening 3D NIfTI Files
In order to demonstrate the procedure, the NIfTI data files "avg152T1_LR_nifti.nii.gz" (https://nifti.nimh.nih.gov/nifti-1/data) and "avg152T1_RL_nifti.nii.gz" (https://nifti.nimh.nih.gov/nifti-1/data/avg152T1_RL_nifti.nii.gz) have been loaded using the Open NIfTI item in the File menu. These data sets are useful to test proper interpretation of coordinate systems since they contain explicit labels for left and right (see below).
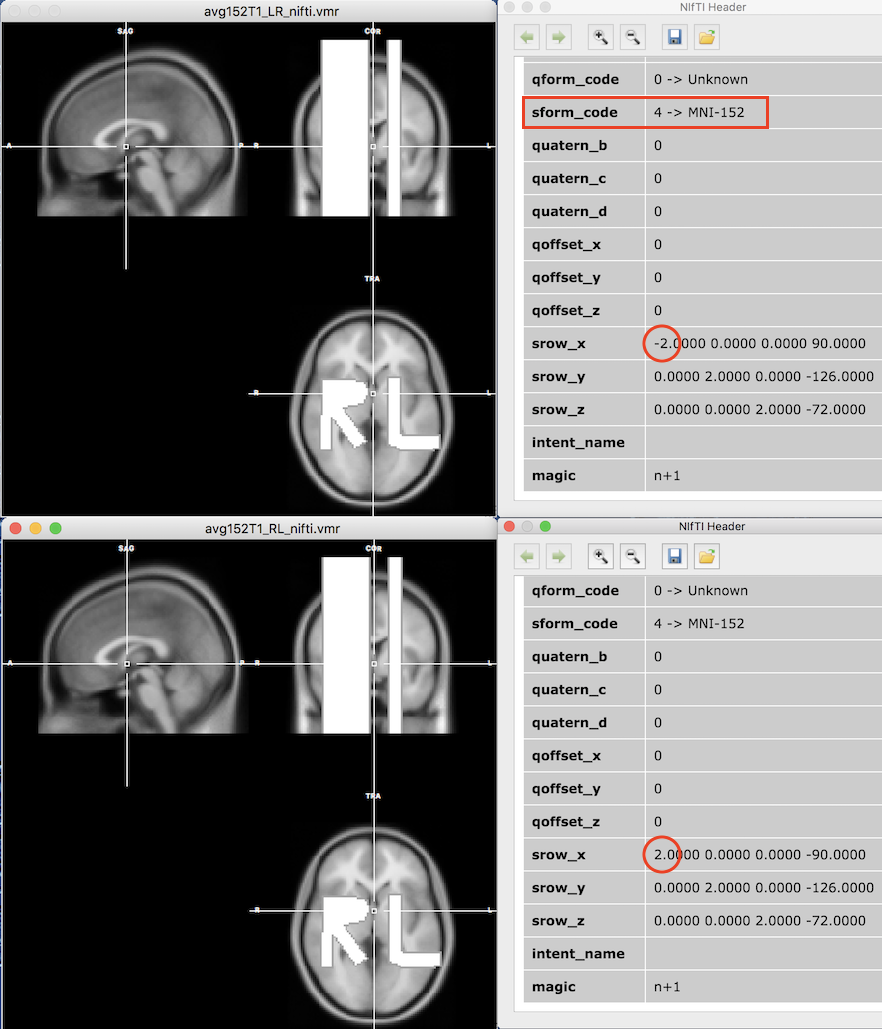
After loading the NIfTI files, the two data sets appear as standard VMR files in BV's multi-document area together with the extracted header information displayed in an internal HTML viewer. As can be seen on the left side in the figure above, both volumes look the same despite the fact that the "_LR_" NIfTI file stores the x axis in opposite direction (radiological convention, right-to-left) than the "_RL_" file (neurological convention, left-to-right). The sform transformation matrix stored in the header ensures that the direction of the x axis is inverted for the "_LR_" file (see marked value -2.0 in the "_LR_" header in the snapshot above) but not for the "_RL_ file (see marked value 2.0 in the "_RL_" header) ensuring that both data sets are transformed to the same RAS+ space. The values of 2.0 in the diagonal of the sform transformation matrix also ensure that the volume in the file is scaled from voxels with a 2mm iso-voxel size to 1 mm MNI-152 space. The translation values in the 4th column of the matrix are used to set the origin (x=0, y=0, z=0) of the MNI RAS+ coordinate system.
If BrainVoyager would use internally a RAS+ system, the application of the sform matrix would be sufficent to interpret the MNI (and Talairach) space NIfTI data. Since BrainVoyager uses internally a PIL coordinate system, a final RAS-to-BV transformation matrix is applied to the data, which reorients the volume using (multiples of) 90 degree rotations and eventually axis flips that do not require data resampling. The NIfTIvoxel-to-RAS and RAS-to-BV matrices are multiplied together so that the whole transformation can be performed in one step using the calculated NIfTIvoxel-to-BV matrix. For the actual transformation, this matrix is finally inverted since BrainVoyager iterates over voxel coordinates in the target (e.g. 256 x 256 x 256) volume and accesses the corresponding values in the original NIftI volume "backwards".
NIfTI Settings
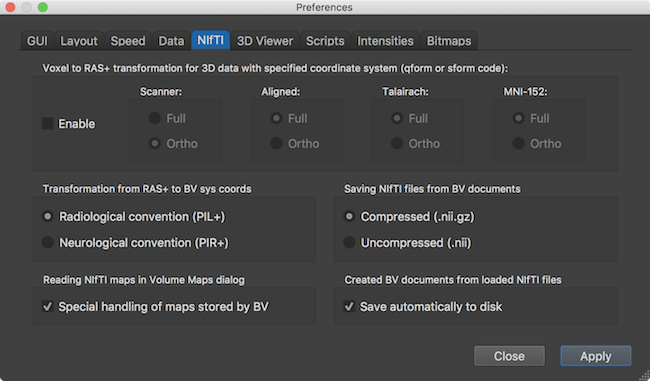
While the transformed data is shown in radiological (right-to-left) convention as default, this can be switched to neurological (left-to-right) convention using the respective options in the Transformation from RAS+ to BV sys coords field in the NIfTI tab of the Preferences (macOS) or Settings (Windows, Linux) dialog (see screenshot above). The NIfTI tab can also be used to specify for which target reference space a full transformation (application of the sform (qform) NIfTIvoxel-to-RAS matrix) should be applied (default for Aligned, Talairach and MNI space) or whether only a "reduced" otho-reorientation matrix should be used (default for Scanner space). In order to read the data without any spatial transformation, the Enable option can be turned off (not recommended). Note that in this case the orientation of a loaded NIfTI file will probably not fit to the sagittal, coronal and transversal view planes.
When loading NIfTI volume map files from the Volume Maps dialog, they are treated in a special way in case that they were saved by BrainVoyager (e.g. avoiding resampling of maps with lower relative resolution with respect to the hosting anatomy). If one wants to load a BrainVoyager stored NIfTI map like a map from other neuroimaging software, the Special handling of maps stored by BV option can be turned off in the Reading NIfT maps in Volume Maps dialog field.
As default, a created native BrainVoyager document (e.g. VMR, FMR-STC, VMR-VTC) is saved to disk after opening a 3D or 4D NIfTI file. In case this is not desired, the Save automatically to disk option in the Created BV documents field can be unchecked. If not auo-saved, the created document can, of course, still be saved to disk using the standard Save Document icon in the main toolbar or the Save item in the File menu. The Preferences / Settings dialog can also be used to specify whether documents saved as NIfTI files should be saved uncompressed or compressed (default) by either checking the Compressed (.nii.gz) or Uncompressed (.nii) option in the Saving NIfTI files from BV documents field. Saving compressed NIfTI files saves space on disk but works substantially slower for large (e.g. 4D time course) files than saving uncompressed files.
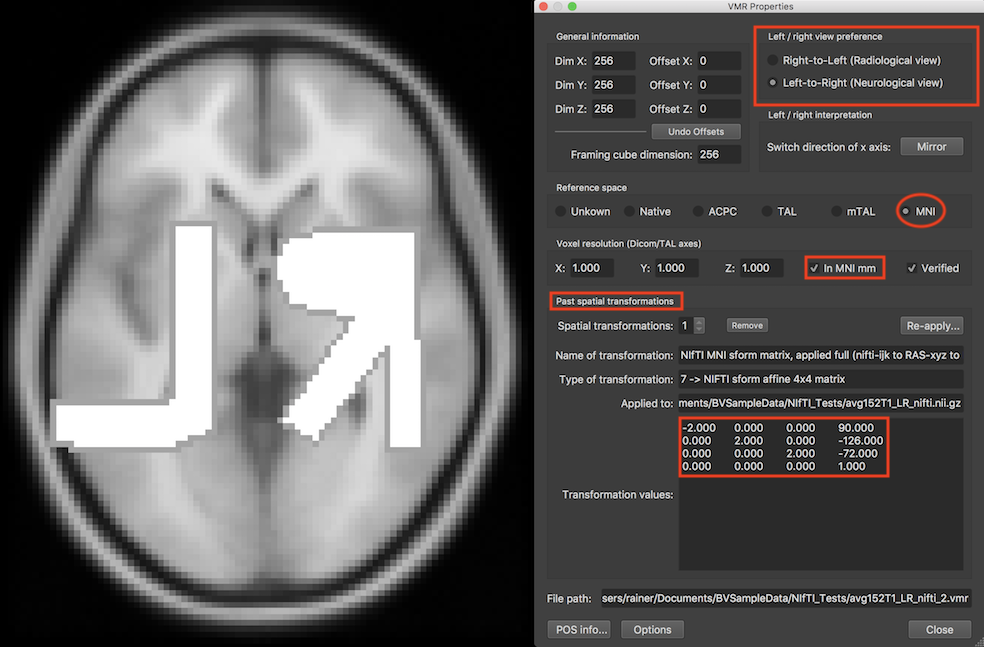
The left/right convention can also be changed afterwards in the Left / right view preference field of the VMR Properties dialog (see screenshot above). The dialog also indicates that the created VMR file is in MNI space using standard MNI mm coordinates. Furthermore, the dialog shows that the VMR header keeps the original affine sform (or qform) transformation matrix in the Past spatial transformations field.
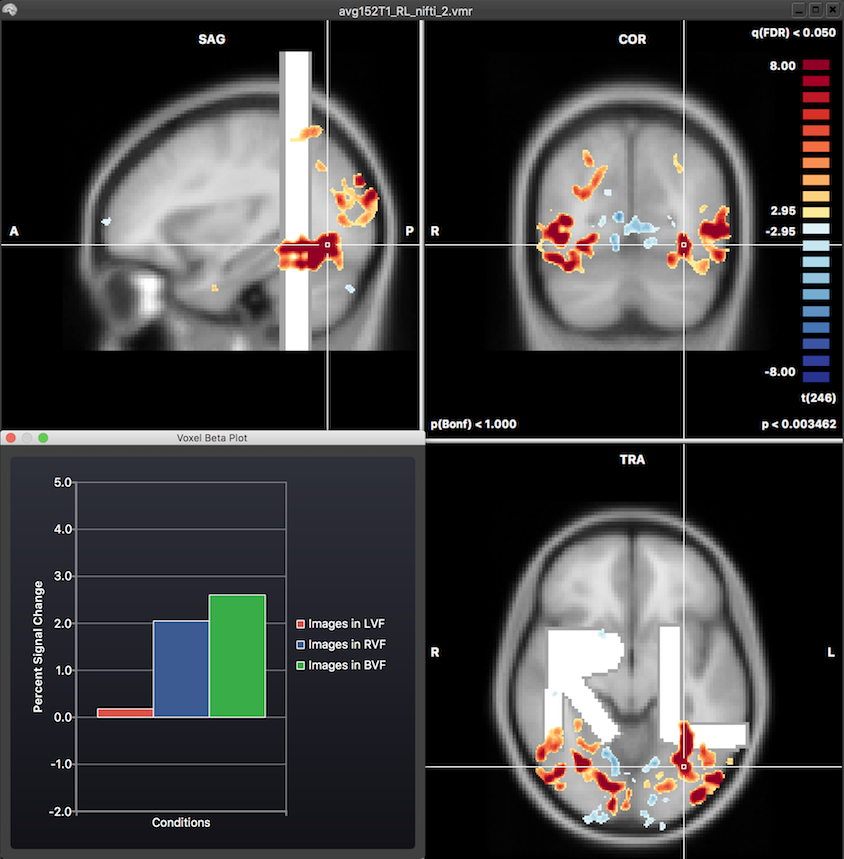
A major goal of the described NIfTI-to-BV transformation steps is to allow using NIfTI data in the same way as when it would have been created within BrainVoyager. While not possible in all cases, most scenarios are already possible (see also topic NIfTI Volume Map Files). When one opens, for example, a NIfTI MNI-152 or Talairach space anatomical data set, it can be directly used as the hosting VMR for statistical analyses in normalized space using VTC files normalized to the same space. In the snapshot below, the created VMR file after loading the "avg152T1_LR_nifti.nii.gz" NIfTI file has been used to link a MNI VTC file that has been created in BrainVoyager from the "Objects" sample data followd by a standard GLM analysis.
FMR-VMR Coregistration
Since the NIfTI transformation matrix is stored in both FMR and VMR files, BrainVoyager's standard header-based initial alignment (IA) step can be performed in the same way as when the FMR / VMR data were created directly from header data of the original scanner (e.g. DICOM) data. For FMR-VMR coregistration, the FMR and VMR files need to be created from NIfTI files containing original data measured in the same scanning session. When starting the header-based initial alignment from the 3D Volume Tools dialog of a VMR file (see snapshto below), the respective qform / sform matrices stored in the respective headers is extracted and used as described for NIfTI header-based FMR-VMR alignment.
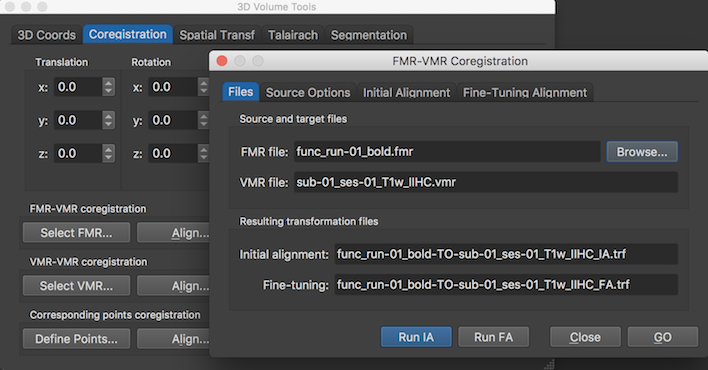
The screenshot below shows the result after running the FMR-VMR header-based alignment for the two example data sets opened from example same-session NIfTI files. The obtained result is very similar to the one obtained when performing the alignment from FMR / VMR files created from the original DICOM files.
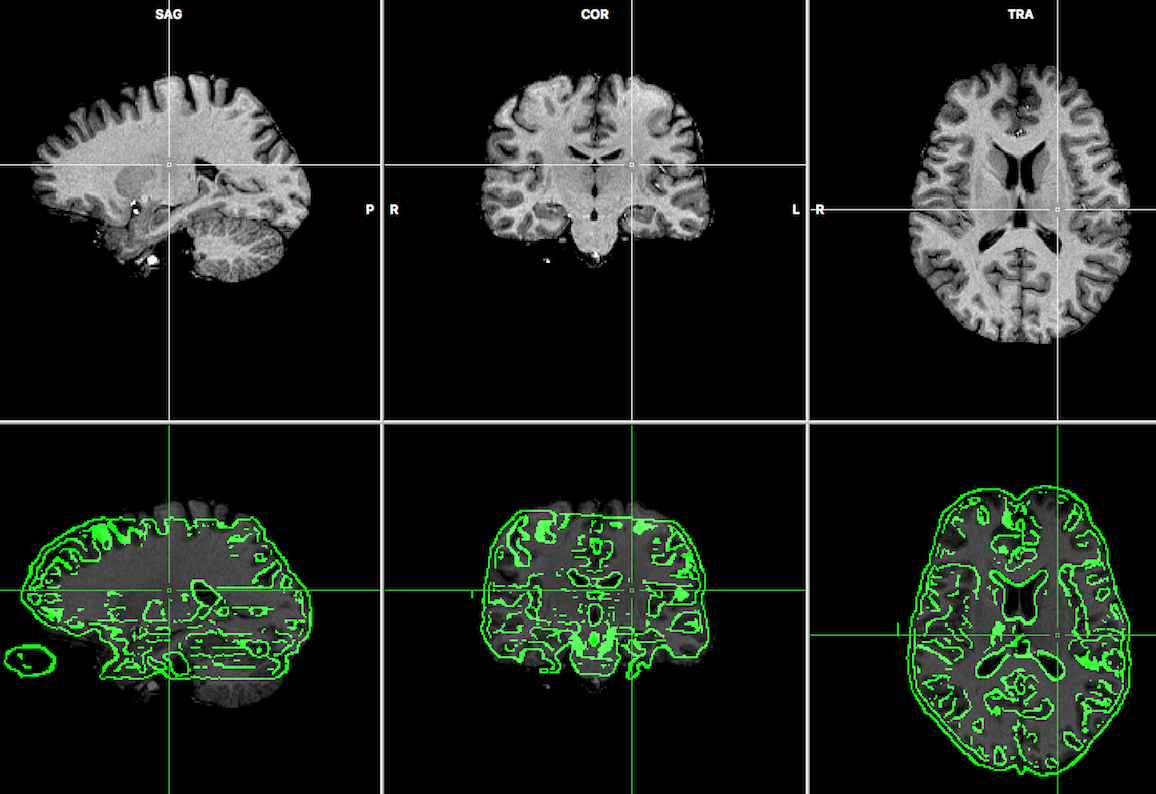
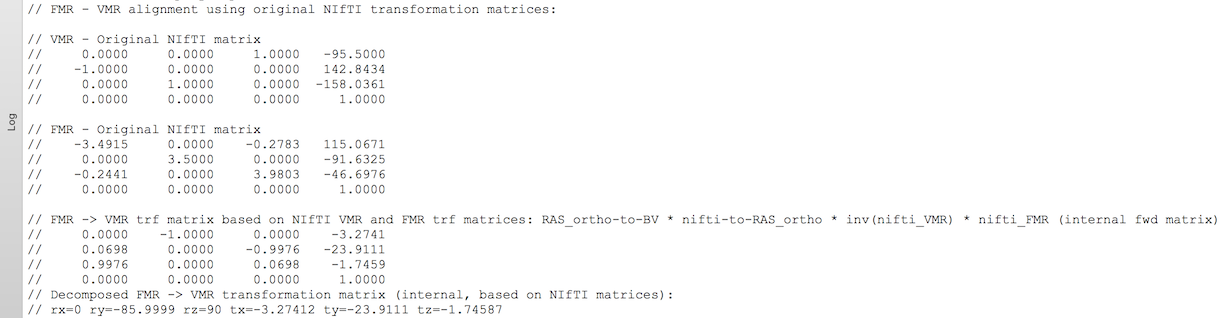
Information about the NIfTI header-based FMR-VMR alignment is also printed in the Log pane (see snapshot below) showing the original NIfTI transformation matrices and the calculated BrainVoyager initial alignment matrix that is also stored automatically to disk for subsequent VTC creation. Note that the FMR-VMR IA matrix stores only orientation and position (of volume center) information, i.e. scaling data is extracted from the involved FMR and VMR data sets when actually performing the initial (header-based) alignment.
After successful FMR-VMR alignment, the standard fine-tuning adjustment step can be performed as usual using either the gradient-based or boundary-based registration appraoch. Note that when the NIfTI files were created by BrainVoyager together with JSON sidecar files, BrainVoyager may perform the IA alignment step using the original DICOM data in case it is found in the associated .json file.
Saving NIfTI Files
An open 3D VMR/V16 or 4D FMR-STC document in native reference space can be converted into a NIfTI file by using the Save NIfTI item in the File menu. Running this function will create and fill a NIfTI heder with appropriate information from the respective BrainVoyager document. In case of VMR documents it is at present assumed that they have been created from original (preferentially DICOM) data so that information to create a "Scanner" coordinate system can be constructed; in this case the sform_code value of the NIfTI header is set to "1" and the sform matrix is filled with the transformation matrix. The matrix is constructed using original DICOM header "RowDir" and "ColDir" orientation vectors; furthermore the center of the first and last slice DICOM location stored in the VMR header is transformed to build a transformation vector for the first voxel in the file with indices i=0,j=0,k=0. Finally any transformations (To-Sagittal orientation, iso-voxelation etc.) are applied inverted to the current VMR before saving the data to disk.
Note. In case of a VMR document, it is checked whether a .V16 file with a matching base file name exists - if a V16 is available, its 2-byte data will be stored in the NIfTI file, otherwise the 1-byte data of the VMR will be saved, and the NIfTI header "datatype" field will be adjusted accordingly.
Besides the NIfTI file, a JSON sidecar file will be saved containing the "BrainVoyagerInfo" object with additional information, including a list of past spatial transformations. The JSON sidecar file will be used by BrainVoyager when loading the NIfTI file in order to convert the anatomical or functional data to the same state when saved as a NIfTI file. In case that a protocol is attached to a functional (FMR) document, it will be stored both inside the JSON file and additionally as a tab-spearated value (TSV) file. Note that the "BV-docs-to-NIfTI conversion" operates similar to the "DICOM-to-NIfTI conversion" described in topic NIfTI with JSON and TSV Sidecars except that the documents stored with the Save NIfTI function may contain not only original data but also (pre-)processed data.
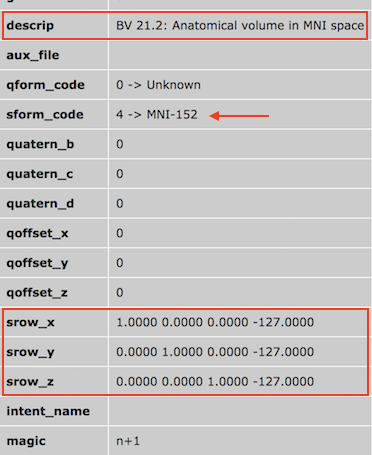
VMR files in normalized reference space (MNI, Talairach) can also be saved to disk. In this case the sform transformation matrix will be set to a matrix without rotation. The created NIfTI header used the description string to identify BrainVoyager as the generator of the file together with information about the type and reference space of the data (see screenshot above).
Current Limitations. Saving documents as NIfTI is currently supported for (preprocessed, e.g. inhomgeneity corrected) 3D anatomical (VMR) data and 4D functional (FMR) data in native and normalized space. Furthermore, volume map data.can be loaded from NIfTI and saved as NIfTI. Support for diffusion-weighted data (DMR, VMR-VDW documents) will be added in an upcoming release.
Copyright © 2023 Rainer Goebel. All rights reserved.