BrainVoyager v23.0
Measuring Cortical Thickness in Volume Space
Cortical thickness measurement requires as input a structural (VMR) file, which has been successfully segmented using the advanced segmentation tools or (preferred) the deep neural network segmentation tool. The result of the advanced segmentation procedure is a file with a name like "[core-name]_WM_GM.vmr" containing both the WM / GM boundary and the GM / CSF (pial) boundary. After loading this file, you can launch the Cortical Thickness Measurement dialog (see below) by clicking the Cortical Thickness Measurement item in the Volumes menu.
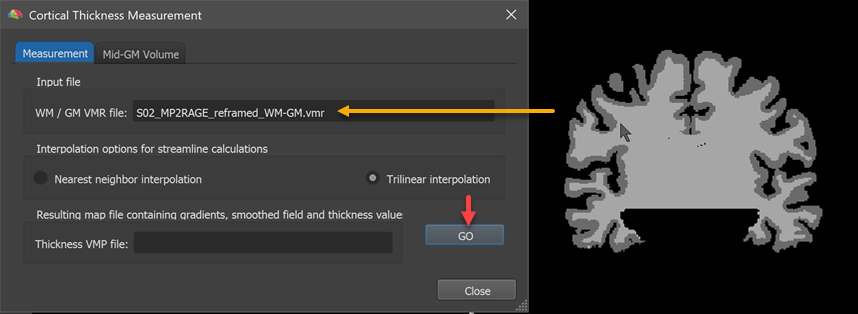
The name of the loaded VMR file with the WM / GM and GM / CSF boundaries will appear in the WM / GM VMR file text box (see orange arrow in the screenshot above). The Interpolation options for streamline calculations field allows to select either the Nearest neighbor interpolation or Trilinear interpolation (recommended) option for the computation of "streamlines" (see below). To measure cortical thickness, simply click the GO button. The result will be a volume map (VMP) file containing submaps with the cortical thickness estimates as well as other useful information. The name of the resulting VMP file stored to disk is displayed in the Thickness VMP file text box.
Note. In case one has used sub-millimeter data for laminar and columnar fMRI, the resulting "[name]_WM-GM_Thickness.vmp" file is all what is needed and one can proceed, for example, with whole cortex or local cortical depth analysis.
As an optional step, the program offers the possibility to calculate a volume and a VOI with a border running approximately through the middle of grey matter. With the calculated cortical thickness information (see below), such calculations can now be perfromed. Note that the mid-level boundary can be used for cortex reconstruction, which might be preferred by some users instead of the WM / GM boundary.
When clicking the GO button in the Measurement tab, the following computations are performed that are based on the Laplace method:
- Three tissue classes are identified in the VMR file with respect to a voxel's intensity value i: CSF (i < 75), GM (75 ≤ i ≤ 125) and WM (i > 125 ). Before starting the main computations, these identified tissue classes are set to three intensity values: CSF: 50, GM: 100, WM: 150. The CSF and WM intensity values are not changed during subsequent computations ("border voltages"), only the intensities of GM voxels are updated incrementally. The starting intensity values are converted to real numbers (floats) in order to allow fine-grained calculations. The resulting volumes are stored as float volume maps (VMPs).
- The intensities of GM voxels are iteratively smoothed (200 iterations) in order to solve Laplace's equation. The resulting smoothed intensities are stored as map 4 ("Laplace smoothed") in the resulting VMP file (see figure below).
- At each voxel, gradient vectors are computed. Each gradient component (x, y, z) is saved in an individual map within the VMP file (maps 1 - 3, see figure below). While the gradient vector lenghts are normalized to 1.0 internally, the component values are saved multiplied by 10.0 in order to simplify thresholding for visualization purposes.
- For every GM voxel, a streamline is calculated using a small step size of 0.1. From a starting GM voxel, the gradient is first followed in one direction (i.e. "up") and then in the opposite direction (i.e. "down"). The integrated values of the accumulated step sizes until a boundary voxel is reached results in a path length. The pathlengths from the two directions are finally added together to obtain the thickness measure for the GM voxel. The computed thickness measures for all GM voxels are stored in map 5 ("Cortical Thickness") in the resulting VMP (see figure below). From the same information a final "WM Distance" map (map 6) is calculated that stores how far a grey matter voxel is away from the white matter boundary.
After these calculations have been completed, the cortical thickness map (map 5) is shown superimposed on the VMR data. The VMP data structure resides in working memory but has been also saved to disk for later usage, especially for cortical thickness analysis in cortex space. Note that this file is very large (about 450 MB). If you click on Overlay Maps in the Analysis menu, the Volume Maps dialog appears with similar entries as shown in the snapshot below.
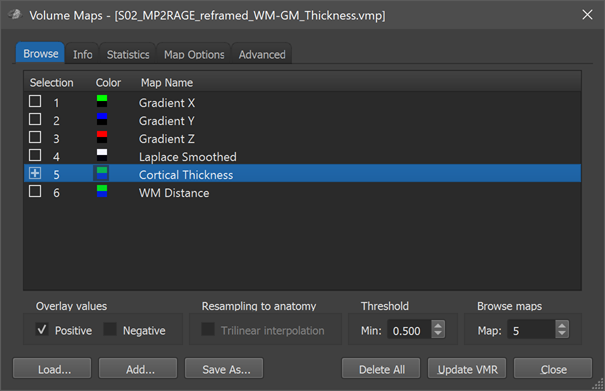
When opening the Volume Maps dialog, the statistical look-up table assigned to the "Cortical Thickness" map is shown, which may better represent the differences between thickness values as compared to the standard overlay colors. The program automatically loads the "Thickness.olt" look-up table from the folder "MapLUTs", which should be in your BrainVoyager installation folder. This look-up table shows thin cortex in blue and thick cortex in green. Since the computed maps have been saved to disk, you may superimpose the thickness map (or any other of the 6 maps) also on other coregistered VMR data sets of the same subject, for example the original, unsegmented data.
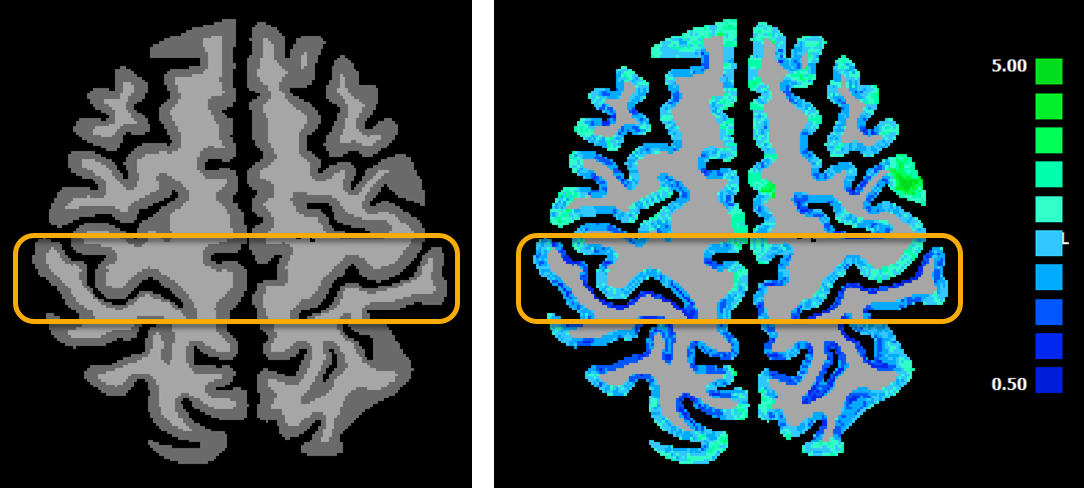
The figure above shows on the left side the input data set (result from a DNN segmentation) and on the right side the resulting cortical thickness map for a selected axial slice. The orange box highlights regions around the central sulcus. It is known that the posterior bank of the central sulcus has a much thinner cortical thickness than the anterior bank. While this can be readily seen already in the segmented data (left side), the cortical thickness map (right side) quantifies this effect and the resulting values (anterior bank ca. 3.1, posterior bank ca. 1.6) are in accordance with the values reported in the literature. Since the central sulcus thickness values in the anterior and posterior bank differ substantially in individual brains, the values obtained there should be used as a first check whether the segmentation and thickness measurement procedures have worked satisfactorily. The effect is easily visible in the obtained cortical thickness map since the posterior bank contains mostly dark blue colors while the anterior bank contains mostly light blue and light green colors. The scale bar on the right shows the mapping from the colors to measured cortical thickness in millimeter.
Note: You can "probe" the cortical thickness measured at each voxel by moving the mouse across voxels - also in the Zoom View pane - and observing the thickness values in the displayed voxel info tooltip.
Copyright © 2023 Rainer Goebel. All rights reserved.