BrainVoyager v23.0
From Volume to Mesh Time Courses
BrainVoyager supports (statistical) analysis of functional MRI data in surface space in the same way as in volume space allowing e.g. to calculate surface maps (SMPs) on folded, inflated and flattened cortex meshes from time series that are attached to the vertices of a mesh. This allows more advanced cortical (statistical) analyses than sampling volume maps - calculated in volume space - from meshes to create surface maps (see topic Overlaying Surface Maps on Cortex Meshes). Besides analysis and visualization of individual time course data on cortex meshes, the creation of mesh time courses also allows advanced group studies by using cortex-based (curvature driven) alignment. In order to attach time series data to mesh vertices, the respective (cortex) mesh can sample the data available in a volume time course (VTC) file resulting in mesh time course (MTC) data. In order to work correctly, the mesh and the VTC data need to be in the same space, i.e. the cortex mesh (usually a single hemisphere) need to have been reconstructed from the brain in the associated VMR document window.
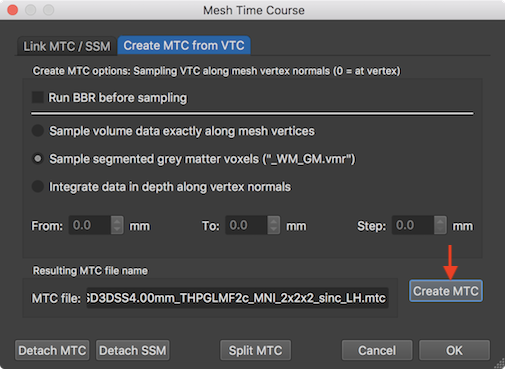
After loading a VMR and after linking the volume time course (VTC) file, a reconstructed (hemisphere) mesh needs to be loaded that will be used to sample the VTC data. A mesh time course (MTC) file can be created using the Mesh Time Courses dialog that can be invoked by clicking the Mesh Time Course item in the Meshes menu. The Create MTC from VTC tab of the Mesh Time Courses dialog contains the Create MTC button that can be used to start the sampling of the VTC data from each vertex of the loaded (and current) mesh file. If the option Sample volume data exactly along mesh vertices is selected, the mesh vertices are projected into the VTC data and the time series are read at the respective vertex positions. Since the vertex coordinates are real values, they usually look for data at positions that fall in between the voxel grid; the values are then determined by trilinear interpolation.
The Create MTC options: Sampling VTC along mesh vertex normals field offers three approaches how the mesh samples the underlying functional data in the associated volume space. When selecting the Sample volume data exactly along mesh vertices option, no depth integration is actually performed, i.e. the vertices will sample the VTC data from the voxels in volume space that are closest to the loation of the mesh vertices using trilinear interpolation.
The Sample segmented grey matter voxels option and the Integrate in depth along vertex normals opton also sample from voxels close to the location of mesh vertices but values are integrated along the normal of a vertex, i.e. along the direction that is orthogonal to the surface area around a vertex. Since the mesh surface extends tangentially along the cortex, sampling along the vertex normal, thus, integrates data along the depth of the cortex. Since the cortex has a thickness in the range of 2-5 mm, depth integration is relevant even with a standard resolution of 2 mm voxels. The range of sampling along the normal can be specified for the Integrate in depth along vertex normals option. The entry in the From value box specifies where to start the depth sampling process with respect to the coordinates of a vertex, i.e. the default value of -1.0 starts the depth integration 1 mm "backwards" towards white matter. The default starting value is not set to 0.0 since it is assumed that a "RECOSM" mesh is used as default which typically runs not exactly along the white-grey matter boundary but about 1 voxel (usually corresponding to 1 mm) inside grey matter. In case that a cortex mesh is used that runs exatly along the white-grey matter boundary (e.g. a "WM" mesh or a cortex mesh resulting from the advanced segmentation pipeline), a value of 0.0 can be used in that field. From the specified starting point, sampling along the normal proceeds with a step size in millimeter that is specified in the Step value box. The default value for the To field is set to 3.0, i.e. depth integration proceeds up to 3 mm through grey matter towards CSF. The values sampled along the normal of a vertex (e.g. at relative depth values -1.0, 0.0, 1.0, 2.0, 3.0) when using the default settings) are averaged resulting in the final value attached to the vertex for one time point. The whole time course is sampled from the VTC data creating a full time course for the respective vertex.
The Sample segmented grey matter voxels option (added in BrainVoyager 21.0) implements a similar approach than the Integrate in depth along vertex normals option. The difference is that the depth sampling range need not to be specified since it is automatically limited to grey matter voxels. This option thus does not require specifiying the sampling range along the normal and it is particularly useful in light of the fact that the cortex varies in thickness. This option is used as default in case that an associated VMR volume with an explicit labelling of grey matter voxels is available, otherwise the option is disabled. To be enabled, grey matter voxels need to have the intensity value of 100 since that value is used to mask grey matter. Since the automatic advanced segmentation pipeline produces a file with explicit grey and white matter labels (with values 100 and 150 respectively), the option is turned on only if the file name of the associated VMR volume ends with the "_WM_GM.vmr" substring as indicated in brackets at the end of the name of this option. If one does not have such a VMR file, one can also use the region growing tool in the 3D Volume Tools dialog to approximately label grey matter voxels using the "range" tool, saving the resulting VMR file under a name ending with the "_WM_GM.vmr" substring.
Since BrainVoyager 20.2 it is possible to optimize the sampling by first performing a boundary-based registration (BBR) procedure. The Run BBR before sampling option is not turned on as default since it might not be necessary, e.g. in case that BBR has been already used for the alignment of functional and anatomical volume data. To ensure however optimal alignment of the current mesh and the VTC data, enabling the BBR option is recommended. If enabled, BBR is performed before VTC sampling by creating a target VMR from the first (functional) VTC volume, followed by running BBR to find and apply the spatial transformation to align the mesh to the target volume. Then the VTC data is sampled on the mesh vertices as described above; finally the location of mesh vertices in space that were adjusted by BBR are put back to the original values. Besides standard BBR output in the Log tab and saved transformation files, this process runs without GUI changes.
After repeating the sampling process for all mesh vertices, the complete mesh time course data has been created and will be saved under the name shown (and eventually modified) in the MTC file text field of the Resulting MTC file name field. The resulting MTC file is automatically linked to the mesh but this can also be done explicitly using the Browse button in the Mesh time course (MTC) file field in the Link MTC / SSM tab of the dialog (or from the title bar context menu of the 3D Viewer). Note that the time courses at vertices can be visualized in teh same way as for voxels in volume space by CTRL-clicking (CMD-clicking on Mac) on any location of the mesh. Single-run statistical GLM analyses can be launched to calculate and visualize surface maps.
Copyright © 2023 Rainer Goebel. All rights reserved.