BrainVoyager v23.0
From FMR-STCs to VTCs in Talairach Space
FMR-STC data can be transformed to normalized (Talairach) space by using the Create VTC dialog (see below) that can be invoked using the Create Volume Time Course (VTC) Data item in the Analysis menu. The menu item is only enabled in case that a VMR project is selected as the current document in the document workspace. The VMR document that is selected when calling the Create VTC dialog should be the high-resolution (usually 1 mm) intra-session (native) 3D anatomical data set.
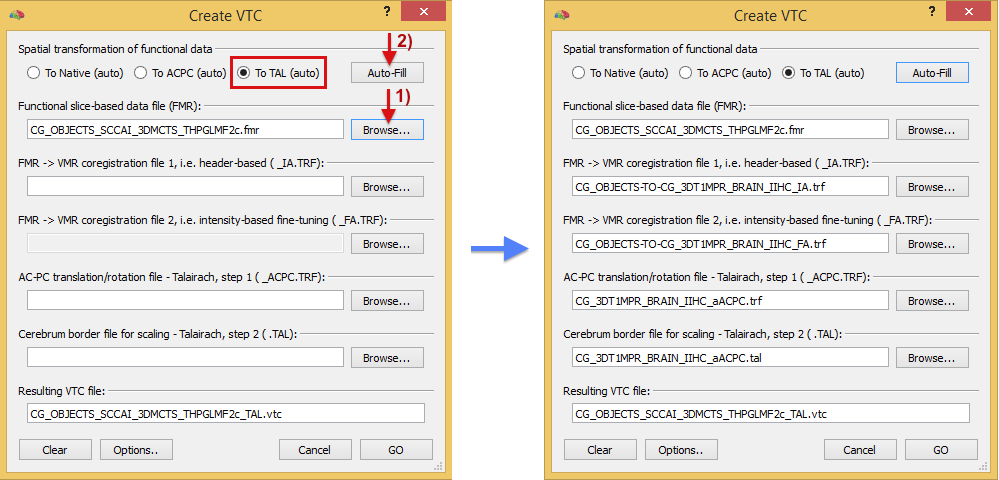
The Create VTC dialog expects the source FMR-STC file that will be transformed into a VTC file as well as several spatial transformation files. The Browse button next to the Functional slice-based data file (FMR) entry can be used to select the source FMR file. In the snapshot above, the .FMR file "CG_OBJECTS_SCCAI_3DMCTS_THPGLMF2c.fmr" has been selected as the source for VTC creation. The dialog allows to specify transformation files that bring the FMR-STC data into native, ACPC and normalized (Talairach) space. If all files are available on disk for Talairach space transformation, the To TAL option is automatically selected in the Spatial transformation of functional data field. If this option is not selected automatically, the To TAL option needs to be selected, which will enable all 4 entries for transformation files in the dialog that are necessary for Talairach transformation. In case that the transformation files for a target space (native, ACPC, Talairach) are available on disk, the options in the Spatial transformation of functional data field will be marked with the "auto" text (e.g. To TAL (auto)) indicating that all open slots can be filled automatically by clicking the Auto-Fill button (see button marked by arrow 2). The right side above shows the automatically selected transformation files for the example case after running the auto-fill routine. In case that the program can not localize the necessary files (e.g. if they are named in an unexpected way or if they are located in different folders), the 4 spatial transformation files can be manually selected using the respective Browse buttons. The first two transformation files are the initial and fine-tuning .trf files as obtained by the FMR-VMR coregistration procedure., i.e. the FMR-VMR coregistration file 1 is the result of header-based intra-session coregistration and the FMR-VMR coregistration file 2 is the result from the fine-tuning adjustment coregistration step. With these files (or at least the first one), the FMR-STC data would be brought into the (native) space of the intra-session 3D anatomical measurement. In order to transform the data to normalized Talairach space, the AC-PC transformation/rotation file - Talairach, step 1 .trf file and the Cerebrum border file for scaling - Talairach, step 2 .tal file need to be provided as obtained by anatomical (automatic) Talairach transformation of the intra-session VMR data set. The name of the resulting VTC file can be entered in the Resulting VTC file text field, which contains as default a suggested file name that is based on the name of the provided FMR file to which the target space ("_TAL" in the example above) is appended folled by the ".vtc" extension. When all files are provided, the transformation process can be started by clicking the GO button.
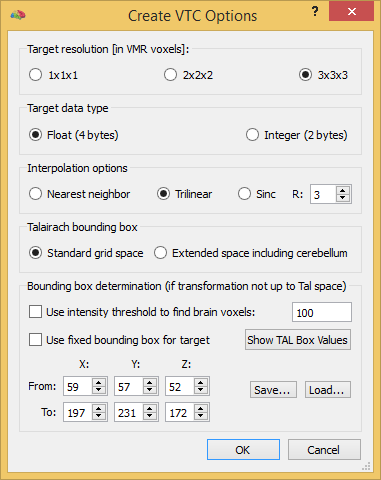
When running the VTC creation procedure, some additional default settings are used that can be modified in the Create VTC Options dialog that can be invoked by clicking the Options button in the Create VTC dialog prior to clicking the GO button. The first field, Target resolution [in VMR voxels], is used to specify the relative resolution of the created VTC data and the hosting VMR data set. The default setting of 3x3x3 will create a VTC where each VTC voxel corresponds to a cube of 27 VMR voxels, i.e. the resolution of the VTC data is 3x lower in each dimentsion than the resolution of the VMR data. This default setting is appropriate for 1 mm anatomical VMR data and functional data with a resolution of about 3 mm (or lower) in each dimension as is typical for standard fMRI experiments. In case of functional data with higher spatisl resolution, the 2x2x2 or 1x1x1 option should be selected. Note that for subsequent statistical group analyses, the same relative resolution option should be used for all data sets included in the study. Note that the target resolution is expressed relative to the hosting VMR data set; since VMR data has usually a iso-voxel resolution of 1 millimeter, the relative values can be usually interpreded as (Talairach) millimeter. There are, howeer, cases such as in the context of high-resolution imaging, where the resolution of the VMR data set is higher (or lower) than 1 millimeter. If, for example, a VMR data set has a resolution of 0.6 millimeter and a 2x2x2 target resolution is selected, the resulting VTC will have a resolution of 1.2 x 1.2 x 1.2 (Talairach) millimeter.
The Target data type field can be used to specify whether the intensity values at each voxel are stored in 4 byte float values (default) or in 2 byte short integers. The Float (4 bytes) option is recommended and should be usually used since float values more precisely represents resampled (preprocessed, saptially transformed) values; the Integer (2 bytes) option should only be used if disk/memory space is limited since the resulting VTC file requires only half the disk/memory space.
When transforming FMR-STC data to Talairach space, the resulting VTC data will be in a common space for each subject that is restricted to a standard sub-volume covering the anterior-poster and left-right extent of the brain; it also covers the superior-inferior extent of the cortex but not the whole brain since the inferior border of the Talairach volume is located at the lowest point of the cortex. In case that a researcher is interested that the cerebellum is included the Extended space including cerebellum option should be selected instead of the default Standard grid space option. The options in the Bounding box determination option are described in the VTC Data in Native and ACPC Space topic.
Schematic Summary of the Talairach VTC Creation Process
The image below summarizes the Talairach space VTC creation procedure. On the left side, the source FMR-STC document is shown containing voxel time courses in measurement (slice) space. The VMR document in the middle shows the same-session (also called intra-session) 1 millimeter 3D anatomical data set as measured in the scanner; the original anatomical measurement space is also called "native" because no spatial transformations have been applied (except iso-voxel and to-sagittal reorientation, if necessary). During the FMR-VMR coregistration process, the FMR slices are brought into the native 3D space. In order to represent anatomical and functional data in Talairach space, the native space VMR is submitted to the brain normalization procedure.
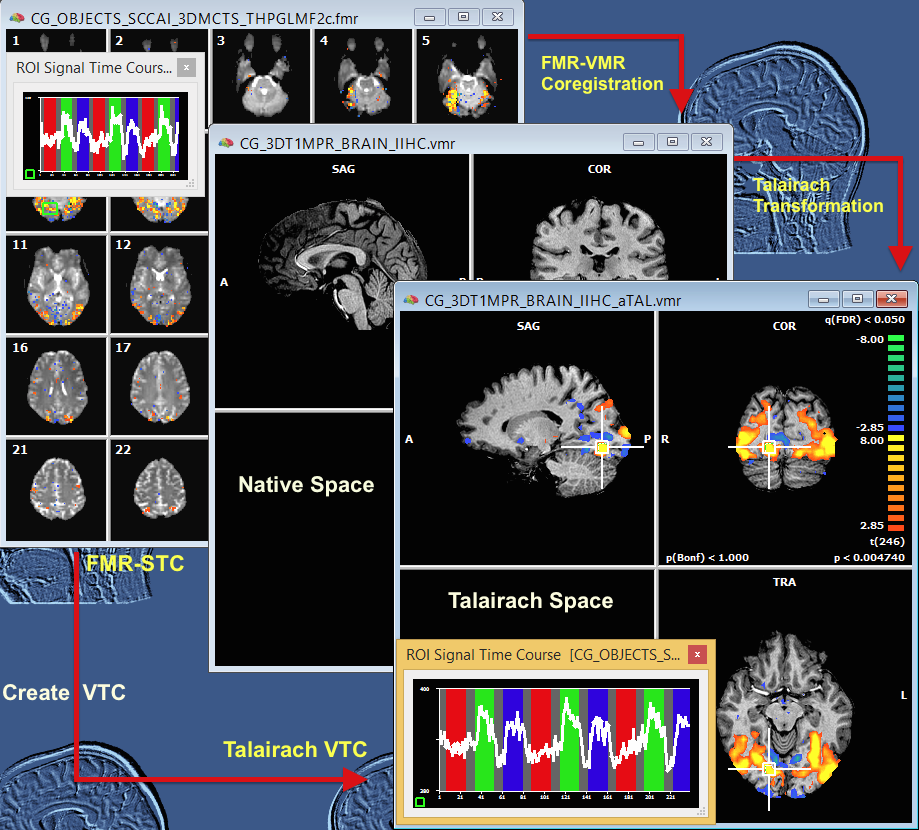
The resulting Talairach space VMR is shown in the document on the right side. The FMR-VMR coregistration and anatomical Talarach procedure provide the necessary transformation files (automatically saved to disk) to convert the slice-based FMR-STC data into Talairach 3D VTC data (indicated on the left lower part of the image). The resulting Talairach VTC file has been linked to the anatomical Talairach VMR file in the document depicted on the right side using the Link Volume Time Course (VTC) dialog that can be invoked with the Link Volume Time Course (VTC) File item in the Analysis menu. Control-clicking on a anatomical voxel will now show the corresponding voxel time course from the Talairach space VTC data. Statistical (GLM) analyses can now be performed in Talairach space (in addition to FMR-STC space) as indicated by the calculated overlayed statistical volume map on the right. Note that by transforming the FMR time course data from different subjects into a common (Talairach/MNI) space, whole-brain (statistical) group analyses can be performed.
Copyright © 2023 Rainer Goebel. All rights reserved.