BrainVoyager v23.0
From DICOM Files to Documents
BrainVoyager supports the most common DICOM files when creating documents in BrainVoyager formats (e.g. VMR, FMR, DMR) and when converting DICOM to NIfTI files. Processing of DICOM files is complex since scanner manufacturers use different strategies to store the data of a single document as a DICOM series over one or more files to disk. Besides different multi-image storage strategies, DICOM file headers are complex, used differently by different manufacturers, and require a rather sophisticated and time-consuming parsing process. Since only some essential information is crucially needed in the context of neuroimaging, the much simpler NIfTI format has been developed with a short header that can be quickly parsed.
Originally MRI DICOM files could only store one image per file. While the standard was improved to allow multiple images per file, most manufacturers continued to use the one-image-per-file approach for a long time. A typical 1mm iso-voxel anatomical T1 scan is often stored as a series of about 200 individual files.
Renaming DICOM Files
DICOM files might be structured on disk in different ways. In one approach, all DICOM files belonging to one scanning session are stored in a single folder. In other scenarios, each series (scan) is stored in a seperate sub-folder. It also happens that DICOM files belonging to the same series are spread over sub-folders. Before creating documents from DICOM files, BrainVoyager first renames the files in one or more folders with the intention that the files become better human-readable and also alphanumerically sortable. The files obtained from a scanner typically look like the following (only a few files shown):
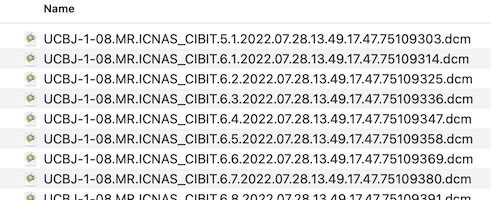
After renaming, the first files in the folder look like this:
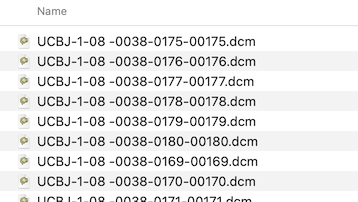
Each header is inspected and the values of the important DICOM tags Series Number, Acquisition Number and Instance Number are extracted and used to rename the files in a folder:
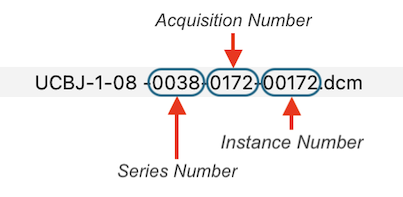
After renaming, DICOM files belonging together are then easily identified as those that have the same series number. While the series number identifies a scan, the acquisition number usually identifies a volume within a functional or diffusion-weighted scan. The instance number usually corresponds to individual slices in the one-image-per-file approach or it replicates the volume index (acquisition number) in case that all slices of one volume are stored in a single DICOM file (see below). BrainVoyager renames files automatically when creating documents but the renaming routine can also be called explicitly using the Rename DICOM Files (see screenshot below) or Anonymize DICOM Files dialogs that can be launched from the File menu.
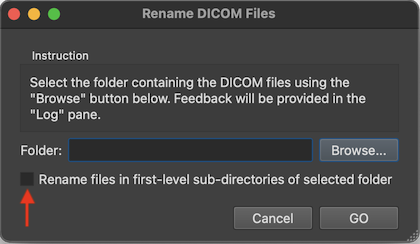
To rename the files in a specific folder, one can select it using the Browse button on the right side of the Folder field. If one wants to rename the DICOM files in all sub-folders of a folder, one can turn on the Rename files in first-level sub-directories of selected folder option (introduced in BV 22.4). One then selects the parent folder that contains the DICOM folders one wants to rename.
DICOM Series Overview Table
There are many possibilities to create BrainVoyager documents from DICOM files or to convert DICOM to NIfTI files including the Create Document Wizard, the Create Document dialog, the Data Analysis Manager and by calling document creation commands in notebooks or scripts. When using the Create Document Wizard as described in subsequent topics, one can browse to a folder with the desired DICOM files and select any file in that folder (see screenshot below).
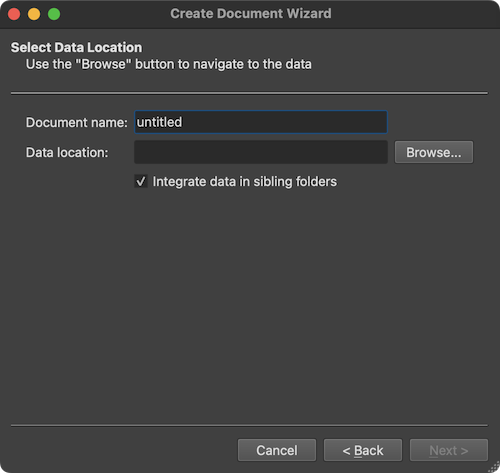
The screenshot above shows the Select Data Location page of the wizard with the option Integrate data in sibling folders (introduced in BV 22.4). If this option is turned on (default), not only the DICOM files in the selected folder but also DICOM files in any sibling folder will be analyzed (and renamed if needed). If the option is turned off, only the DICOM files in the selected folder will be analyzed. After clicking the Browse button and the selection of a DICOM file in a folder, the program presents the DICOM Series Overview dialog presenting all scans (series) in a table that were found in the selected as well as in all sibling folders (if the option is checked):
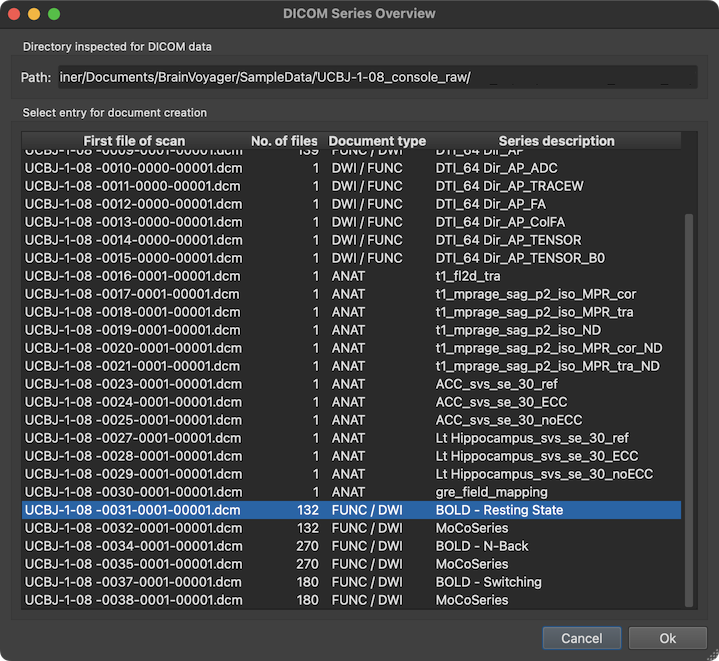
The screenshot above shows an example with a DICOM series overview table integrating data from multiple sibling folders. The series (scan) to which the selected file belongs is highlighted automatically but also any other series can be selected. The column First file of scan of the series overview table shows the first file of each series (in the example series '31' is selected). The No. of files columns shows how many DICOM files belonging to the series have been detected on disk. Based on the number of files and the DICOM variant (see below), the Document type column provides a guess which document type a series likely represents (in the example, the selected series is assumed to be a functional or diffusion-weighted scan). The Series description column shows the name of the series extracted from the DICOM header. In the selected series above, the name "BOLD - Resting State" identifies the series as representing a functional resting state scan.
DICOM Header
After clicking the OK button in the DICOM Series Overview dialog, the program returns to the Create Document Wizard (left side in screenshot below) and presents the DICOM header of the first file of the selected DICOM series in a DICOM Header dialog (see right side of the screenshot below).
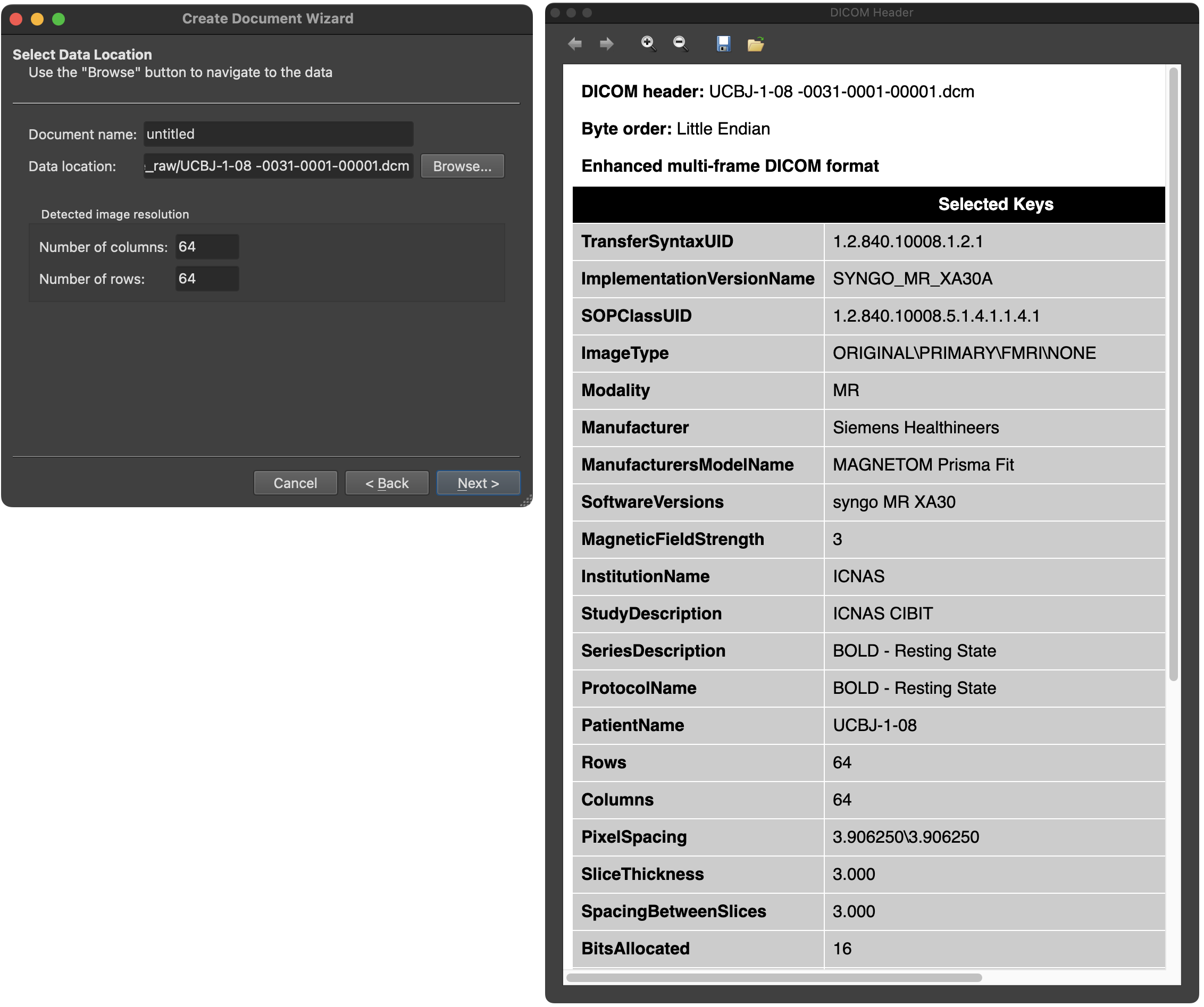
The DICOM header shows a non-complete list of DICOM keys and their respective values. Above the header, additional information about the DICOM file is shown, including its name, the byte order and whether the file is an enhanced multi-frame DICOM file containing multiple images (see below).
Single-Image DICOM Files
As mentioned at the begin of this topic, DICOM files originally did only store a single image per file and this 'single-image-per-file' approach is still in use today. The single-image files of a DICOM series are read by BrainVoyager and integrated into a single document file. For a 3D anatomical scan with 190 slices, for exsample, the program extracts the image data from each file and 'stacks' them together to form a VMR document or 3D NIfTI file. The 'one imager per file' approach gets, however, inconvenient when considering functional MRI scans that consists of many repeated measurements of the same slices over time requiring thousands of DICOM files. A typical functional scan with, for example, 200 volumes of 50 slices would result in 10,000 single-image DICOM files. Thus, reading the data of one functional run of one subject already requires parsing the header and extracting the image data of 10,000 files.
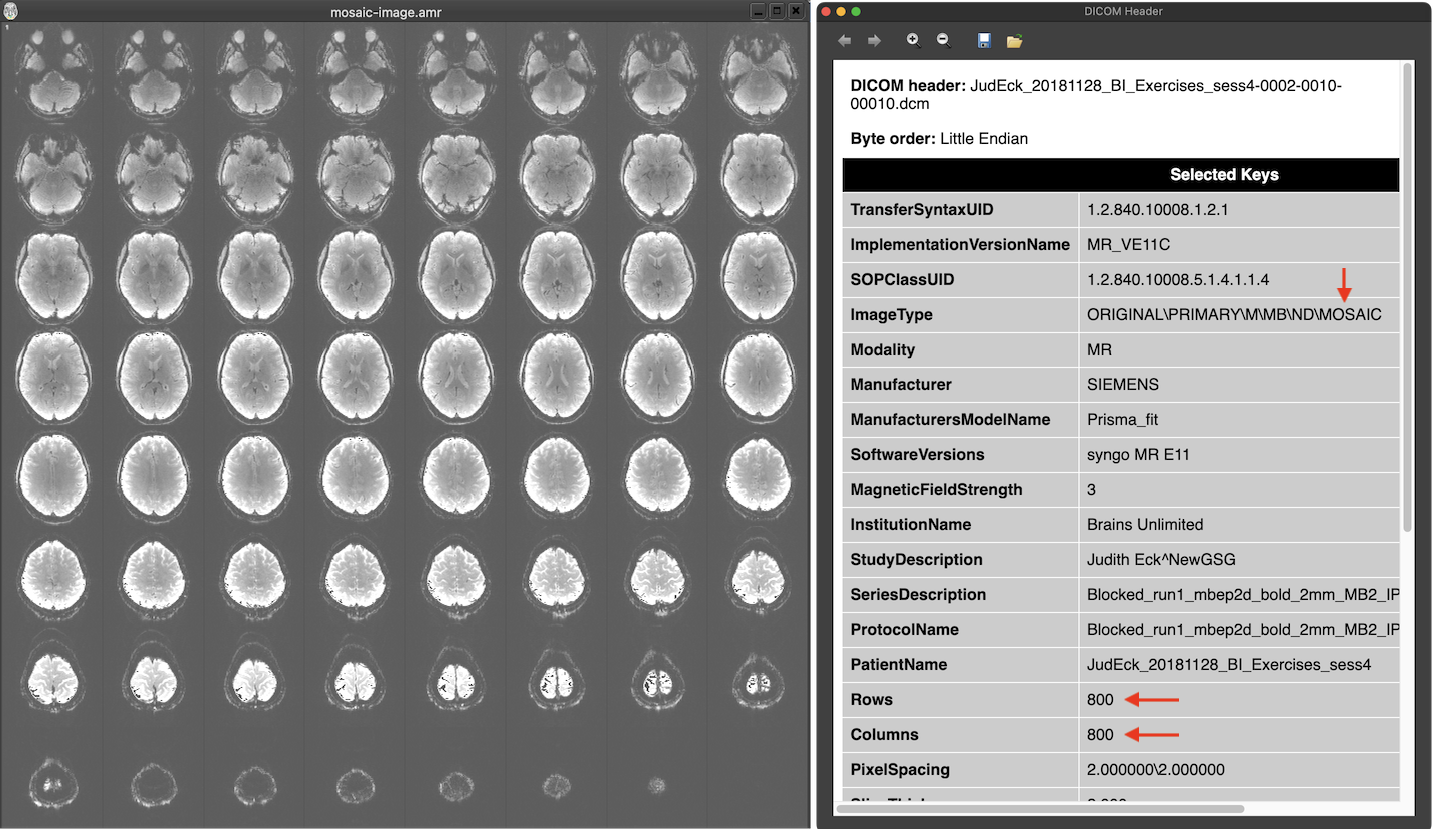
Manufacturers circumvented this issue by either avoiding DICOMs using custom formats (such as PAR/REC for Philips scanners) or by packing multiple images in one file without violating the 'one image per file' principle. The latter approach has been implemented by Siemens using the 'mosaic' format that packs multiple slices in a single (large) image. In the screenshot above the text in the ImageType DICOM field of the header contains the string 'MOSAIC' revealing that this is a moasic file. The Rows and Columns fields describe a large image with 800 rows and 800 columns in this pseudo 'one image per file' DICOM file. When looking on the image extracted from the DICOM file (left side above), one sees that the image consists actually of 8x8 small (100 x 100) slices that make up one volume of the functional scan. The smaller images need to be extracted from the mosaic image to be able to process them as separate slices usually forming the slices of a single functional or diffusion-weighted volume. Since DICOM allows to add custom tags in the header, information about the contained actual slice images (e.g. rows and columns, slice timing information) is described in manufacturer-specific fields. This approach reduces the number of required DICOM files to the number of volumes (200 in the example above) instead of thousands of files. The enhanced DICOM standard (see below) provides a standard solution to store a series of 'frames' (images) in a single DICOM file.
Enhanced Multi-Frame DICOM Files
While already around for some time, the enhanced DICOM 'multipe images per file' format was only slowly adopted by manufacturers, especially Siemens. With their new 'XA' software platform (that seems to be also brought to older scanners running the VB or VE platforms), Siemens now also suports the enhanced DICOM format. The enhanced DICOM format allows to store and describe in the header any number of frames, which are usually the slices of functional and diffusion-weighted volumes. The multiple images (potentially even multiple stacks of slices with different orientation) described in the header are stored in a sequence of 'frames' at the end of the file (in the 'Pixel Data' DICOM tag). The header contains general tags describing the stored data as a whole as well as per-frame data (in sequence tags), which contain, for example, the position and slice acquisition information for each frame (image).
Note that not only the slices of functional and diffusion volumes are stored in a single enhanced DICOM file but also the slices of an anatomical dataset can be stored in a single multi-frame DICOM file. As for functional volumes, position (and orientation) information for each slice is stored in the enhanced DICOM header.
Copyright © 2023 Rainer Goebel. All rights reserved.