BrainVoyager v23.0
Trial Estimation
In order to obtain as many training exemplars as possible for each class, response values are extracted or estimated for each individual trial as opposed to estimating a voxel's mean response of a condition in a standard GLM. Estimated single-trial responses across relevant voxels (e.g. from a region-of-interest) then form the feature vectors used to train the classifier. The estimated trial response might be as simple as the activity level at a certain time point (e.g. at the time of the expected hemodynamic peak response) or the mean response of a few measurement points around the peak response relative to a pre-stimulus baseline. More often, however, a GLM is used to fit an expected hemodynamic response to the measured trial data. The beta value estimating the amplitude of the hemodynamic response is then used as the trial response value. For a more stable fit of the model, additional confound predictors (next to the constant) may be added, such as a predictor for estimating a linear trend. Instead of estimated beta values, t values derived from the beta values may be chosen as output; since t values take the variability of the estimated parameter into account, they usually provide more reliable results.
Application
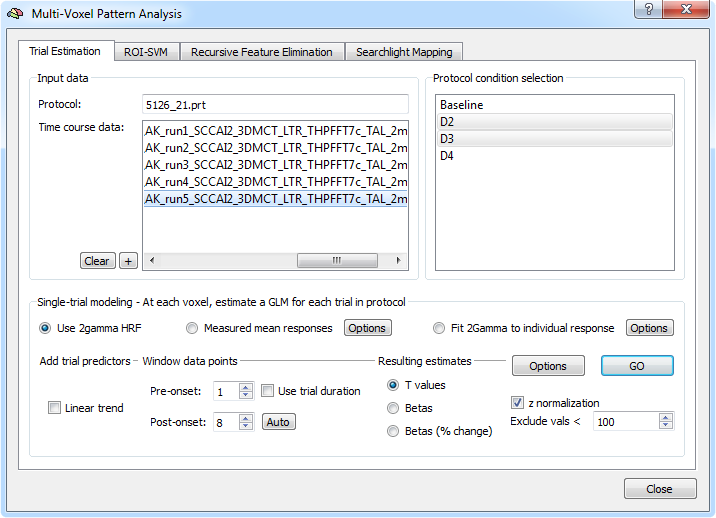
Trial estimation can be performed using the Trial Estimation tab of the Multi-Voxel Pattern Analysis (MVPA) dialog. Using the "+" button in the Input data field, the time course data (one VTC file for each included run) of the respective experiment can be selected (note that multi-file selection can be used). The selected data will be listed in the Time course data box in the Input data field (see snapshot above). From a selected functional data file, the referenced protocol is shown in the Protocol text field and the associated experimental conditions are extracted and displayed in the Protocol condition selection box for the currently selected file. For the example data, the conditions displayed are "Baseline", "D2", "D3" and "D4" (the "D's" represent digits of the stimulated hand). The first two main conditions are selected automatically as targets for single-trial estimation (the first condition is skipped under the assumption that the first condition is "baseline" or "rest"). It may be useful to select additional conditions (e.g. "D4" in this example) in case that multiple pair-wise classifications (e.g. "D2" vs "D3", "D2" vs "D4", "D3" vs "D4") or multi-class comparisions are planned.
GLM-Based Trial Extraction
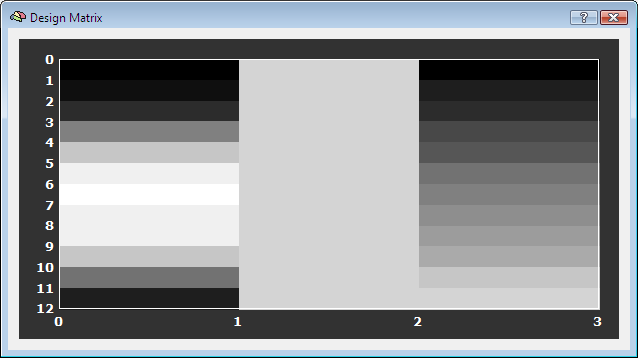
For estimating beta or t values per trial, a two-gamma HRF is used as default (the Use 2gamma HRF option is turned on). The Fit 2Gamma to individual response option is a more flexible approach fitting the onset in a specified interval of the two-gamma function and selecting the response shape leading to the best GLM fit (use the corresponding Options button to modify the default search options). The Measured mean responses option can be used to use empirically measured fMRI responses for fitting single-trial GLMs (use the corresponding Options button for detailed settings). When the Linear trend option is turned on in the Add trial predictors field, a linear confound predictor is added to the design matrix next to the main and constant predictors (see snapshot below).
The trial estimation step calculates a GLM for each trial per voxel by creating a data window around trial onset. The data window for a trial is specified with respect to the onset of a trial as specified in the protocol(s) of the included functional data file(s). The Pre-onset spin box can be used to specify how many data points should be included before the trial onset data point. The offset of the trial data window can be either specified absolutely or with respect to the duration of the trial as specified in the protocol. If the Use trial duration option is turned off, the end of the trial window is specified with the Post-onset spin box specifying the number of time points included after the stimulus onset point. If, for example, the pre-onset value is 2 and the post-onset value is 8, the data window w used for estimating the trial response would be w = pre-onset + 1 + post-onset = 11 data points. Clicking the Auto button after setting the pre-onset interval will provide a value for the post-onset interval, ensuring that the interval is just long enough to let the expected BOLD response turn back (largely) to baseline. The described way to specify the trial data window is well-suited in case that all trials have the same duration. In case that trials of a condition may have different durations, the trial duration option should be used to specify the data window. If the Use trial duration option is turned on, the offset is determined by the trial duration plus the value specified in the Post-trial spin box as w = pre-onset + trial-duration + post-trial; in case of a pre onset value of 2, a post-trial value of 4 and a trial duration of 5 this would, for example, result in a data window of w = 2 + 5 + 4 = 11 data points. Note that for both versions available to specify the trial data window, the duration of a trial as specified in the protocol will be used to determine a box-car reference function that is set to 1 at the trial onset data point and subsequent points reflecting the trial duration, e.g. 0 0 1 1 1 1 1 0 0 0 0 in the described example case. The constructed box-car reference is finally submitted to the hemodynamic response function to calculate the shape of the predictor as used in the single-trial design matrix.
The options in the Resulting estimates field allow to specify the resulting type of single-trial estimate, which can be t values (T-values option), beta values (Betas option) or percent signal change beta values (Betas (% change) option). As default, the time course data within each trial window is z normalized, but this can be turned off by un-checking the z normalization option. In order to exclude voxels in the background, usually having low signal values, use the Exclude vals < spin box; to turn this voxel selection filter off, change the value to "0".
Note: At present, only protocols in "Volumes" resolution are supported. You may convert any protocol in "millisecond" resolution to a protocol in volumes resolution using the Protocol dialog. To adjust for eventually introduced temporal shifts (within a window of one TR), the Fit 2gamma to individual response option should be used when estimating trial responses.

After clicking the GO button, the trial estimates are calculated and stored in volume map (VMP) files. For each included time course (VTC) file, one corresponding VMP output file will be created. These VMP files serve as input for the ROI-SVM analysis. The program will also show the created design matrix used for the windowed GLM trial analysis (see snapshot below). Furthermore, it is useful to consult the Log pane showing additional information.
The naming of the output files contains the "_Trials" substring at the end indicating that the GLM-based extraction method has been used to create this file; furthermore the names of the selected conditions (if together not longer than 100 characters) will appear in the file name. A substring "GLM-" will appear to indicate which of three variants have been used; the string "GLM-2G" indicates that a standard 2-Gamma function has been used to create the trial predictor, the string "GLM-Fit2G" indicates that the 2-Gamma function has been shifted in time for each trial to find the strongest response, and the string "GLM-MeasResp" indicates that a measured (empirical) response is used as the trial's basis function. Futhermore, the file name contains the window in which the response is estimated as well as which parameter (beta, t) value has been created. A typical file name looks like this:
RunA_TAL_motor_imagery-mental_singing_GLM-2G_PreOn-1-PostOn-8_z-t_Trials.vmp
The "RunA_TAL" substring comes from the name of the processed functional run (RunA_TAL.vtc), the substrings "motor_imagery" and "mental_singing" are the selected condition names (in the example 2 out of 3 available conditions); the "GLM-2G" string indicates that a standard 2-Gamma function has been used; the "PreOn-1-PostOn-8 indicates that the interval from -1 to 8 relative to trial onset (0) has been specified; if the Use trial duration option is used, the end of the interval would be indicated by the "PostCond" string. In case that the Linear Trend option would have been turned on, the substring "LT" would have been added to indicate that a linear trend predictor has been included in the single-trial design matrix. The "z-t" substring indicates that t values have been exported and that the trial time course was z-transformed; if beta values are exported, the file name would contain the "b" substring, and a "p" substring if the trial time course would have been percent signal change transformed. This elaborate file naming scheme has the advantage that the file name indicates what options have been used to create it; furthermore, files created with different options for the same data do not overwrite each other.
The created VMP files containing the trial estimates can be submitted to support vector machine classifiers to analyze multivariate differences in specified ROIs. Background information about support vector machines (SVMs) can be found in the section Support Vector Machines.
Raw Data Trial Extraction
As an alternative to the GLM-based (recommended) trial extraction approach, trial responses can also be obtained directly from a voxel's time course by specifying, with respect to trial onset, a single value or the mean of some consecutive values as the trial response. To use this approach, click the Extract Raw button. The appearing Extract Trial Data dialog allows to specify the begin and end point of the extraction interval relative to trial onset in the Extract mean value in interval (0 = trial onset) field. The default value of "3" for both the From and the To fields would select a single value 3 time points after trial onset (0), which would be appropriate for a TR of about 2 seconds. It is important to adjust the extraction interval values with respect of the TR of your study! With a TR of 1 second, for example, a value of 5 or 6 would be appropriate for the extraction interval. With short volume TR values, it may be better to specify a range of values since the hemodynamic peak/plateau will be spread out across several time points, even for short events. In case of a TR value of 1 second, for example, an extraction interval from 4 -7 might be appropriate. If more than one time point is specified, the mean of all values within the interval will be calculated and used as the trial's estimated value.
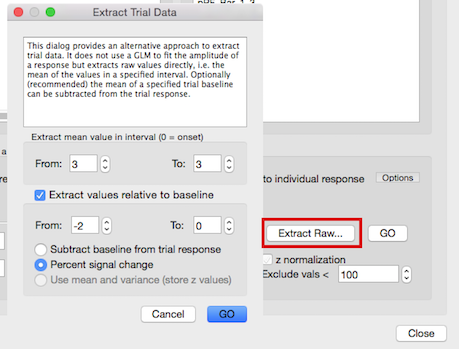
Usually the raw values are not ideal for trial extraction since they might fluctuate around a different signal level in different runs. One approach to handle this issue is to use time course (e.g. VTC) files that have been standardized (e.g. with percent signal change or z normalization of voxel time courses). If, however, standard "raw" data is used (i.e. preprocessed but without time course normalization), the Extract Trial Data dialog offers two approaches to obtain more robust trial data by relating the extracted raw data values to a specified baseline period. To use one of these options, the Extract values relative to baseline field needs to be turned on (default). The interval for calculating the baseline value can be specified in this field using the respective From (default value "-2") and To (default value "0") spin boxes. In case that the Percent signal change option is turned on (default), the trial value (single value or mean of several values) is divided by the baseline to provide a percent signal change value. As an alternative, the calculated baseline value can be simply subtracted from the trial response value by turning on the Subtract baseline from trial response option.
After clicking the GO button, the trials are extracted for each voxel and stored in a series of VMP files (one for each included run). The naming of the output files contains the "_RawDataTrials" substring at the end to indicate that the non-GLM extraction method has been used to create this file; furthermore the chosen extraction interval as well as the names of the selected conditions (if together not longer than 100 characters) will appear in the file name. In case that one of the two baseline options have been used, the specified interval is also stored in the file name; if a percent signal change calculation has been performed, the substring "psc" is added. A typical file name looks like this:
RunA_TAL_motor_imagery-mental_calculation_From-3-To-4_BLFrom--2-BLTo-0_psc_RawDataTrials.vmp
The "RunA_TAL" substring comes from the name of the processed functional run (RunA_TAL.vtc), the substrings "motor_imagery" and "mental_calculation" are the selected condition names (in the example 2 out of 3 available conditions); the "From-3-TO-4" substring indicates the extraction interval (two time points 3-4); the substring "BLFrom--2-BLTo-0" indicates that the extracted values were processed relative to the baseline interval from -2 to 0, and the "psc" substring indicates that the percent signal change option has been used. This elaborate file naming scheme has the advantage that the name indicates what options have been used to create it; furthermore, files created with different options for the same data do not overwrite each other.
The created VMP files containing the trial estimates can be submitted to support vector machine classifiers to analyze multivariate differences in specified ROIs. Background information about support vector machines (SVMs) can be found in the section Support Vector Machines.
Copyright © 2023 Rainer Goebel. All rights reserved.